Boosting the concordance index for survival data--a unified framework to derive and evaluate biomarker combinations
- PMID: 24400093
- PMCID: PMC3882229
- DOI: 10.1371/journal.pone.0084483
Boosting the concordance index for survival data--a unified framework to derive and evaluate biomarker combinations
Abstract
The development of molecular signatures for the prediction of time-to-event outcomes is a methodologically challenging task in bioinformatics and biostatistics. Although there are numerous approaches for the derivation of marker combinations and their evaluation, the underlying methodology often suffers from the problem that different optimization criteria are mixed during the feature selection, estimation and evaluation steps. This might result in marker combinations that are suboptimal regarding the evaluation criterion of interest. To address this issue, we propose a unified framework to derive and evaluate biomarker combinations. Our approach is based on the concordance index for time-to-event data, which is a non-parametric measure to quantify the discriminatory power of a prediction rule. Specifically, we propose a gradient boosting algorithm that results in linear biomarker combinations that are optimal with respect to a smoothed version of the concordance index. We investigate the performance of our algorithm in a large-scale simulation study and in two molecular data sets for the prediction of survival in breast cancer patients. Our numerical results show that the new approach is not only methodologically sound but can also lead to a higher discriminatory power than traditional approaches for the derivation of gene signatures.
Conflict of interest statement
Figures
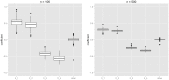
 (left) and
(left) and  (right). Boxplots represent the empirical distribution of the resulting coefficients. Only markers
(right). Boxplots represent the empirical distribution of the resulting coefficients. Only markers  to
to  had an actual effect on the survival time.
had an actual effect on the survival time.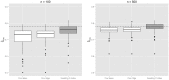
 (left) and
(left) and  (right). Boxplots represent the empirical distribution of the resulting
(right). Boxplots represent the empirical distribution of the resulting  on corresponding test samples. The dotted line corresponds to the discriminatory power resulting from the true combination of predictors with known coefficients.
on corresponding test samples. The dotted line corresponds to the discriminatory power resulting from the true combination of predictors with known coefficients.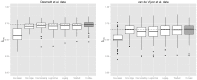
 on corresponding test samples. The dotted line corresponds to the median
on corresponding test samples. The dotted line corresponds to the median  -index resulting from the new approach.
-index resulting from the new approach.Similar articles
-
Boosting the discriminatory power of sparse survival models via optimization of the concordance index and stability selection.BMC Bioinformatics. 2016 Jul 22;17:288. doi: 10.1186/s12859-016-1149-8. BMC Bioinformatics. 2016. PMID: 27444890 Free PMC article.
-
Identifying cancer biomarkers by network-constrained support vector machines.BMC Syst Biol. 2011 Oct 12;5:161. doi: 10.1186/1752-0509-5-161. BMC Syst Biol. 2011. PMID: 21992556 Free PMC article.
-
NCC-AUC: an AUC optimization method to identify multi-biomarker panel for cancer prognosis from genomic and clinical data.Bioinformatics. 2015 Oct 15;31(20):3330-8. doi: 10.1093/bioinformatics/btv374. Epub 2015 Jun 18. Bioinformatics. 2015. PMID: 26092859
-
Predicting censored survival data based on the interactions between meta-dimensional omics data in breast cancer.J Biomed Inform. 2015 Aug;56:220-8. doi: 10.1016/j.jbi.2015.05.019. Epub 2015 Jun 3. J Biomed Inform. 2015. PMID: 26048077 Free PMC article.
-
A gradient boosting algorithm for survival analysis via direct optimization of concordance index.Comput Math Methods Med. 2013;2013:873595. doi: 10.1155/2013/873595. Epub 2013 Nov 20. Comput Math Methods Med. 2013. PMID: 24348746 Free PMC article.
Cited by
-
Development and external validation of prognostic nomograms in hepatocellular carcinoma patients: a population based study.Cancer Manag Res. 2019 Apr 10;11:2691-2708. doi: 10.2147/CMAR.S191287. eCollection 2019. Cancer Manag Res. 2019. PMID: 31118768 Free PMC article.
-
A novel risk score model based on eight genes and a nomogram for predicting overall survival of patients with osteosarcoma.BMC Cancer. 2020 May 24;20(1):456. doi: 10.1186/s12885-020-06741-4. BMC Cancer. 2020. PMID: 32448271 Free PMC article.
-
Liver- and Microbiome-derived Bile Acids Accumulate in Human Breast Tumors and Inhibit Growth and Improve Patient Survival.Clin Cancer Res. 2019 Oct 1;25(19):5972-5983. doi: 10.1158/1078-0432.CCR-19-0094. Epub 2019 Jul 11. Clin Cancer Res. 2019. PMID: 31296531 Free PMC article.
-
A Novel Staging System to Forecast the Cancer-Specific Survival of Patients With Resected Gallbladder Cancer.Front Oncol. 2020 Jul 28;10:1281. doi: 10.3389/fonc.2020.01281. eCollection 2020. Front Oncol. 2020. PMID: 32850391 Free PMC article.
-
Prognosis prediction model based on competing endogenous RNAs for recurrence of colon adenocarcinoma.BMC Cancer. 2020 Oct 7;20(1):968. doi: 10.1186/s12885-020-07163-y. BMC Cancer. 2020. PMID: 33028275 Free PMC article.
References
-
- Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, et al. (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independentvalidation series. Clinical Cancer Research 13: 3207–3214. - PubMed
-
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AAM, et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine 347: 1999–2009. - PubMed
-
- Kok M, Linn SC, Laar RKV, Jansen MPHM, van den Berg TM, et al. (2009) Comparison of gene expression profiles predicting progression in breast cancer patients treated with tamoxifen. Breast Cancer Research and Treatment 13: 275–283. - PubMed
-
- Li H, Gui J (2004) Partial Cox regression analysis for high-dimensional microarray gene expression data. Bioinformatics 20: 208–215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

