Sequences in the terminal protein and reverse transcriptase domains of the hepatitis B virus polymerase contribute to RNA binding and encapsidation
- PMID: 24401091
- PMCID: PMC4090289
- DOI: 10.1111/jvh.12225
Sequences in the terminal protein and reverse transcriptase domains of the hepatitis B virus polymerase contribute to RNA binding and encapsidation
Abstract
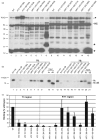
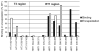
Hepatitis B virus (HBV) antiviral therapy is plagued by limited efficacy and resistance to most nucleos(t)ide analog drugs. We have proposed that the complex RNA binding mechanism of the HBV reverse transcriptase (P) may be a novel target for antivirals. We previously found that RNA binds to the duck HBV (DHBV) P through interactions with the T3 and RT1 motifs in the viral terminal protein and reverse transcriptase domains, respectively. Here, we extended these studies to HBV P. HBV T3 and RT1 synthetic peptides bound RNA in a similar manner as did analogous DHBV peptides. The HBV T3 motif could partially substitute for DHBV T3 during RNA binding and DNA priming by DHBV P, whereas replacing RT1 supported substantial RNA binding but not priming. Substituting both the HBV T3 and RT1 motifs restored near wild-type levels of RNA binding but supported very little priming. Alanine-scanning mutations to the HBV T3 and RT1 motifs blocked HBV ε RNA binding in vitro and pgRNA encapsidation in cells. These data indicate that both the HBV T3 and RT1 motifs contain sequences essential for HBV ε RNA binding and encapsidation of the RNA pregenome, which is similar to their functions in DHBV. Small molecules that bind to T3 and/or RT1 would therefore inhibit encapsidation of the viral RNA and block genomic replication. Such drugs would target a novel viral function and would be good candidates for use in combination with the nucleoside analogs to improve efficacy of antiviral therapy.
Keywords: DNA priming; RNA binding; encapsidation; hepatitis B virus.
© 2014 John Wiley & Sons Ltd.
Figures



References
-
- Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Howley P, Griffin DE, Lamb RA, Martin MA, Roizman B, et al., editors. Fields Virology 5. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2977–3029.
-
- Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: epidemiology and vaccination. EpidemiolRev. 2006;28:112–25. - PubMed
-
- Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. NEnglJ Med. 2004;350(11):1118–29. - PubMed
-
- Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11(2):97–107. - PubMed
-
- Sorrell MF, Belongia EA, Costa J, Gareen IF, Grem JL, Inadomi JM, et al. National Institutes of Health Consensus Development Conference Statement: management of hepatitis B. AnnInternMed. 2009;150(2):104–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

