Tertiary structure prediction and identification of druggable pocket in the cancer biomarker - Osteopontin-c
- PMID: 24401206
- PMCID: PMC3922830
- DOI: 10.1186/2251-6581-13-13
Tertiary structure prediction and identification of druggable pocket in the cancer biomarker - Osteopontin-c
Abstract
Background: Osteopontin (Eta, secreted sialoprotein 1, opn) is secreted from different cell types including cancer cells. Three splice variant forms namely osteopontin-a, osteopontin-b and osteopontin-c have been identified. The main astonishing feature is that osteopontin-c is found to be elevated in almost all types of cancer cells. This was the vital point to consider it for sequence analysis and structure predictions which provide ample chances for prognostic, therapeutic and preventive cancer research.
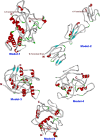
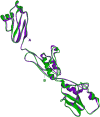
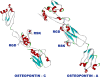
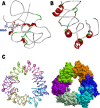
Methods: Osteopontin-c gene sequence was determined from Breast Cancer sample and was translated to protein sequence. It was then analyzed using various software and web tools for binding pockets, docking and druggability analysis. Due to the lack of homological templates, tertiary structure was predicted using ab-initio method server - I-TASSER and was evaluated after refinement using web tools. Refined structure was compared with known bone sialoprotein electron microscopic structure and docked with CD44 for binding analysis and binding pockets were identified for drug designing.
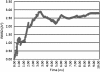
Results: Signal sequence of about sixteen amino acid residues was identified using signal sequence prediction servers. Due to the absence of known structures of similar proteins, three dimensional structure of osteopontin-c was predicted using I-TASSER server. The predicted structure was refined with the help of SUMMA server and was validated using SAVES server. Molecular dynamic analysis was carried out using GROMACS software. The final model was built and was used for docking with CD44. Druggable pockets were identified using pocket energies.
Conclusions: The tertiary structure of osteopontin-c was predicted successfully using the ab-initio method and the predictions showed that osteopontin-c is of fibrous nature comparable to firbronectin. Docking studies showed the significant similarities of QSAET motif in the interaction of CD44 and osteopontins between the normal and splice variant forms of osteopontins and binding pockets analyses revealed several pockets which paved the way to the identification of a druggable pocket.
Figures




Similar articles
-
PockDrug-Server: a new web server for predicting pocket druggability on holo and apo proteins.Nucleic Acids Res. 2015 Jul 1;43(W1):W436-42. doi: 10.1093/nar/gkv462. Epub 2015 May 8. Nucleic Acids Res. 2015. PMID: 25956651 Free PMC article.
-
Ab initio modeling of small proteins by iterative TASSER simulations.BMC Biol. 2007 May 8;5:17. doi: 10.1186/1741-7007-5-17. BMC Biol. 2007. PMID: 17488521 Free PMC article.
-
Integration of QUARK and I-TASSER for Ab Initio Protein Structure Prediction in CASP11.Proteins. 2016 Sep;84 Suppl 1(Suppl 1):76-86. doi: 10.1002/prot.24930. Epub 2015 Sep 23. Proteins. 2016. PMID: 26370505 Free PMC article.
-
Osteopontin and related phosphorylated sialoproteins: effects on mineralization.Ann N Y Acad Sci. 1995 Apr 21;760:249-56. doi: 10.1111/j.1749-6632.1995.tb44635.x. Ann N Y Acad Sci. 1995. PMID: 7785899 Review.
-
Osteopontin.Crit Rev Oral Biol Med. 2000;11(3):279-303. doi: 10.1177/10454411000110030101. Crit Rev Oral Biol Med. 2000. PMID: 11021631 Review.
Cited by
-
Quantitative Analysis of Protein Evolution: The Phylogeny of Osteopontin.Front Genet. 2021 Aug 16;12:700789. doi: 10.3389/fgene.2021.700789. eCollection 2021. Front Genet. 2021. PMID: 34484297 Free PMC article.
-
Sesquiterpene from Polygonum barbatum disrupts mitochondrial membrane potential to induce apoptosis and inhibits metastasis by downregulating matrix metalloproteinase and osteopontin in NCI-H460 cells.Naunyn Schmiedebergs Arch Pharmacol. 2022 Aug;395(8):987-1001. doi: 10.1007/s00210-022-02256-w. Epub 2022 May 23. Naunyn Schmiedebergs Arch Pharmacol. 2022. PMID: 35604429
-
PockDrug-Server: a new web server for predicting pocket druggability on holo and apo proteins.Nucleic Acids Res. 2015 Jul 1;43(W1):W436-42. doi: 10.1093/nar/gkv462. Epub 2015 May 8. Nucleic Acids Res. 2015. PMID: 25956651 Free PMC article.
-
Osteopontin in Vascular Disease.Arterioscler Thromb Vasc Biol. 2019 Apr;39(4):613-622. doi: 10.1161/ATVBAHA.118.311577. Arterioscler Thromb Vasc Biol. 2019. PMID: 30727754 Free PMC article.
-
Osteopontin isoforms differentially promote arteriogenesis in response to ischemia via macrophage accumulation and survival.Lab Invest. 2019 Mar;99(3):331-345. doi: 10.1038/s41374-018-0094-8. Epub 2018 Jun 29. Lab Invest. 2019. PMID: 29959420 Free PMC article.
References
-
- Berns EM, Foekens JA, van Putten WL, van Staveren IL, Portengen H, De Koning WC, Klijn JG. Prognostic factors in human primary breast cancer: comparison of c-myc and HER2/neu amplification. J Steroid Biochem Mol Biol. 1992;43(1–3):13–19. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

