Host-specificity among abundant and rare taxa in the sponge microbiome
- PMID: 24401862
- PMCID: PMC4030224
- DOI: 10.1038/ismej.2013.227
Host-specificity among abundant and rare taxa in the sponge microbiome
Abstract
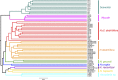
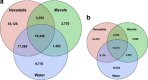
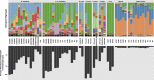
Microbial communities have a key role in the physiology of the sponge host, and it is therefore essential to understand the stability and specificity of sponge-symbiont associations. Host-specific bacterial associations spanning large geographic distance are widely acknowledged in sponges. However, the full spectrum of specificity remains unclear. In particular, it is not known whether closely related sponges host similar or very different microbiota over wide bathymetric and geographic gradients, and whether specific associations extend to the rare members of the sponge microbiome. Using the ultra-deep Illumina sequencing technology, we conducted a comparison of sponge bacterial communities in seven closely related Hexadella species with a well-resolved host phylogeny, as well as of a distantly related sponge Mycale. These samples spanned unprecedentedly large bathymetric (15-960 m) gradients and varying European locations. In addition, this study included a bacterial community analysis of the local background seawater for both Mycale and the widespread deep-sea taxa Hexadella cf. dedritifera. We observed a striking diversity of microbes associated with the sponges, spanning 47 bacterial phyla. The data did not reveal any Hexadella microbiota co-speciation pattern, but confirmed sponge-specific and species-specific host-bacteria associations, even within extremely low abundant taxa. Oligotyping analysis also revealed differential enrichment preferences of closely related Nitrospira members in closely related sponges species. Overall, these results demonstrate highly diverse, remarkably specific and stable sponge-bacteria associations that extend to members of the rare biosphere at a very fine phylogenetic scale, over significant geographic and bathymetric gradients.
Figures


References
-
- Cairns SD. Deep-water corals: an overview with special reference to diversity and distribution of deep-water scleractinian corals. Bull Mar Sci. 2007;81:311–322.
-
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

