Metaproteomic analysis of a winter to spring succession in coastal northwest Atlantic Ocean microbial plankton
- PMID: 24401863
- PMCID: PMC4030229
- DOI: 10.1038/ismej.2013.234
Metaproteomic analysis of a winter to spring succession in coastal northwest Atlantic Ocean microbial plankton
Abstract
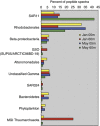
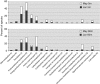
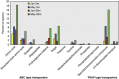
In this study, we used comparative metaproteomics to investigate the metabolic activity of microbial plankton inhabiting a seasonally hypoxic basin in the Northwest Atlantic Ocean (Bedford Basin). From winter to spring, we observed a seasonal increase in high-affinity membrane transport proteins involved in scavenging of organic substrates; Rhodobacterales transporters were strongly associated with the spring phytoplankton bloom, whereas SAR11 transporters were abundant in the underlying waters. A diverse array of transporters for organic compounds were similar to the SAR324 clade, revealing an active heterotrophic lifestyle in coastal waters. Proteins involved in methanol oxidation (from the OM43 clade) and carbon monoxide (from a wide variety of bacteria) were identified throughout Bedford Basin. Metabolic niche partitioning between the SUP05 and ARCTIC96BD-19 clades, which together comprise the Gamma-proteobacterial sulfur oxidizers group was apparent. ARCTIC96BD-19 proteins involved in the transport of organic compounds indicated that in productive coastal waters this lineage tends toward a heterotrophic metabolism. In contrast, the identification of sulfur oxidation proteins from SUP05 indicated the use of reduced sulfur as an energy source in hypoxic bottom water. We identified an abundance of Marine Group I Thaumarchaeota proteins in the hypoxic deep layer, including proteins for nitrification and carbon fixation. No transporters for organic compounds were detected among the thaumarchaeal proteins, suggesting a reliance on autotrophic carbon assimilation. In summary, our analyses revealed the spatiotemporal structure of numerous metabolic activities in the coastal ocean that are central to carbon, nitrogen and sulfur cycling in the sea.
Figures


References
-
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. - PubMed
-
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. - PubMed
-
- Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

