The Open Connectome Project Data Cluster: Scalable Analysis and Vision for High-Throughput Neuroscience
- PMID: 24401992
- PMCID: PMC3881956
- DOI: 10.1145/2484838.2484870
The Open Connectome Project Data Cluster: Scalable Analysis and Vision for High-Throughput Neuroscience
Abstract
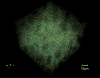
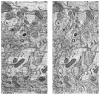
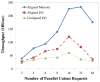
We describe a scalable database cluster for the spatial analysis and annotation of high-throughput brain imaging data, initially for 3-d electron microscopy image stacks, but for time-series and multi-channel data as well. The system was designed primarily for workloads that build connectomes- neural connectivity maps of the brain-using the parallel execution of computer vision algorithms on high-performance compute clusters. These services and open-science data sets are publicly available at openconnecto.me. The system design inherits much from NoSQL scale-out and data-intensive computing architectures. We distribute data to cluster nodes by partitioning a spatial index. We direct I/O to different systems-reads to parallel disk arrays and writes to solid-state storage-to avoid I/O interference and maximize throughput. All programming interfaces are RESTful Web services, which are simple and stateless, improving scalability and usability. We include a performance evaluation of the production system, highlighting the effec-tiveness of spatial data organization.
Keywords: Connectomics; Data-intensive computing.
Figures








References
-
- Abadi DJ, Madden SR, Ferreira M. Integrating compression and execution in column-oriented database systems. SIGMOD. 2006
-
- Baumann P, Dehmel A, Furtado P, Ritsch R, Widmann B. The multidimensional database system RasDaMan. SIGMOD. 1998
-
- Chang C, Acharya A, Sussman A, Saltz J. T2: a customizable parallel database for multidimensional data. SIGMOD Record. 1998;27(1)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
