Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase
- PMID: 24403075
- PMCID: PMC3931030
- DOI: 10.1074/jbc.M113.532226
Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase
Abstract
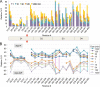
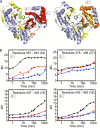
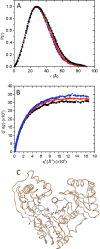
The enzyme phosphomannomutase/phosphoglucomutase (PMM/PGM) from Pseudomonas aeruginosa catalyzes an intramolecular phosphoryl transfer across its phosphosugar substrates, which are precursors in the synthesis of exoproducts involved in bacterial virulence. Previous structural studies of PMM/PGM have established a key role for conformational change in its multistep reaction, which requires a dramatic 180° reorientation of the intermediate within the active site. Here hydrogen-deuterium exchange by mass spectrometry and small angle x-ray scattering were used to probe the conformational flexibility of different forms of PMM/PGM in solution, including its active, phosphorylated state and the unphosphorylated state that occurs transiently during the catalytic cycle. In addition, the effects of ligand binding were assessed through use of a substrate analog. We found that both phosphorylation and binding of ligand produce significant effects on deuterium incorporation. Phosphorylation of the conserved catalytic serine has broad effects on residues in multiple domains and is supported by small angle x-ray scattering data showing that the unphosphorylated enzyme is less compact in solution. The effects of ligand binding are generally manifested near the active site cleft and at a domain interface that is a site of conformational change. These results suggest that dephosphorylation of the enzyme may play two critical functional roles: a direct role in the chemical step of phosphoryl transfer and secondly through propagation of structural flexibility. We propose a model whereby increased enzyme flexibility facilitates the reorientation of the reaction intermediate, coupling changes in structural dynamics with the unique catalytic mechanism of this enzyme.
Keywords: Carbohydrate Biosynthesis; Enzyme Catalysis; Hydrogen-Deuterium Exchange; Phosphorylation; Protein Dynamics; Pseudomonas aeruginosa; Small Angle X-ray Scattering.
Figures
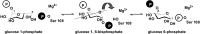

Similar articles
-
The reaction of phosphohexomutase from Pseudomonas aeruginosa: structural insights into a simple processive enzyme.J Biol Chem. 2006 Jun 2;281(22):15564-71. doi: 10.1074/jbc.M600590200. Epub 2006 Apr 4. J Biol Chem. 2006. PMID: 16595672
-
Chemical shift assignments of domain 4 from the phosphohexomutase from Pseudomonas aeruginosa suggest that freeing perturbs its coevolved domain interface.Biomol NMR Assign. 2014 Oct;8(2):329-33. doi: 10.1007/s12104-013-9511-5. Epub 2013 Jul 28. Biomol NMR Assign. 2014. PMID: 23893395 Free PMC article.
-
Phosphorylation in the catalytic cleft stabilizes and attracts domains of a phosphohexomutase.Biophys J. 2015 Jan 20;108(2):325-37. doi: 10.1016/j.bpj.2014.12.003. Biophys J. 2015. PMID: 25606681 Free PMC article.
-
Exopolysaccharide alginate synthesis in Pseudomonas aeruginosa: enzymology and regulation of gene expression.Adv Enzymol Relat Areas Mol Biol. 1995;70:221-55. doi: 10.1002/9780470123164.ch4. Adv Enzymol Relat Areas Mol Biol. 1995. PMID: 8638483 Review. No abstract available.
-
Phosphoryl transfer enzymes and hypervalent phosphorus chemistry.Acc Chem Res. 2004 Oct;37(10):746-53. doi: 10.1021/ar040053b. Acc Chem Res. 2004. PMID: 15491121 Review.
Cited by
-
Mutations in hereditary phosphoglucomutase 1 deficiency map to key regions of enzyme structure and function.J Inherit Metab Dis. 2015 Mar;38(2):243-56. doi: 10.1007/s10545-014-9757-9. Epub 2014 Aug 29. J Inherit Metab Dis. 2015. PMID: 25168163 Review.
-
Biology, Mechanism, and Structure of Enzymes in the α-d-Phosphohexomutase Superfamily.Adv Protein Chem Struct Biol. 2017;109:265-304. doi: 10.1016/bs.apcsb.2017.04.005. Epub 2017 May 17. Adv Protein Chem Struct Biol. 2017. PMID: 28683921 Free PMC article.
-
Multiple Ligand-Bound States of a Phosphohexomutase Revealed by Principal Component Analysis of NMR Peak Shifts.Sci Rep. 2017 Jul 13;7(1):5343. doi: 10.1038/s41598-017-05557-w. Sci Rep. 2017. PMID: 28706231 Free PMC article.
-
Data on the phosphorylation state of the catalytic serine of enzymes in the α-D-phosphohexomutase superfamily.Data Brief. 2016 Dec 15;10:398-405. doi: 10.1016/j.dib.2016.12.017. eCollection 2017 Feb. Data Brief. 2016. PMID: 28050582 Free PMC article.
-
Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding.Biomolecules. 2020 Dec 3;10(12):1631. doi: 10.3390/biom10121631. Biomolecules. 2020. PMID: 33287293 Free PMC article.
References
-
- Smith A. J., Müller R., Toscano M. D., Kast P., Hellinga H. W., Hilvert D., Houk K. N. (2008) Structural reorganization and preorganization in enzyme active sites: comparisons of experimental and theoretically ideal active site geometries in the multistep serine esterase reaction cycle. J. Am. Chem. Soc. 130, 15361–15373 - PMC - PubMed
-
- Hammes-Schiffer S., Benkovic S. J. (2006) Relating protein motion to catalysis. Annu. Rev. Biochem. 75, 519–541 - PubMed
-
- Naught L. E., Tipton P. A. (2005) Formation and reorientation of glucose 1,6-bisphosphate in the PMM/PGM reaction: transient-state kinetic studies. Biochemistry 44, 6831–6836 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

