The potential role of As-sumo-1 in the embryonic diapause process and early embryo development of Artemia sinica
- PMID: 24404204
- PMCID: PMC3880333
- DOI: 10.1371/journal.pone.0085343
The potential role of As-sumo-1 in the embryonic diapause process and early embryo development of Artemia sinica
Abstract
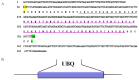
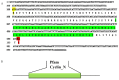
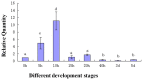
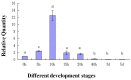
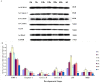
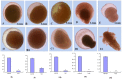
During embryonic development of Artemia sinica, environmental stresses induce the embryo diapause phenomenon, required to resist apoptosis and regulate cell cycle activity. The small ubiquitin-related modifier-1 (SUMO), a reversible post-translational protein modifier, plays an important role in embryo development. SUMO regulates multiple cellular processes, including development and other biological processes. The molecular mechanism of diapause, diapause termination and the role of As-sumo-1 in this processes and in early embryo development of Artemia sinica still remains unknown. In this study, the complete cDNA sequences of the sumo-1 homolog, sumo ligase homolog, caspase-1 homolog and cyclin B homolog from Artemia sinica were cloned. The mRNA expression patterns of As-sumo-1, sumo ligase, caspase-1, cyclin B and the location of As-sumo-1 were investigated. SUMO-1, p53, Mdm2, Caspase-1, Cyclin B and Cyclin E proteins were analyzed during different developmental stages of the embryo of A. sinica. Small interfering RNA (siRNA) was used to verify the function of sumo-1 in A. sinica. The full-length cDNA of As-sumo-1 was 476 bp, encoding a 92 amino acid protein. The As-caspases-1 cDNA was 966 bp, encoding a 245 amino-acid protein. The As-sumo ligase cDNA was 1556 bp encoding, a 343 amino acid protein, and the cyclin B cDNA was 739 bp, encoding a 133 amino acid protein. The expressions of As-sumo-1, As-caspase-1 and As-cyclin B were highest at the 10 h stage of embryonic development, and As-sumo ligase showed its highest expression at 0 h. The expression of As-SUMO-1 showed no tissue or organ specificity. Western blotting showed high expression of As-SUMO-1, p53, Mdm2, Caspase-1, Cyclin B and Cyclin E at the 10 h stage. The siRNA caused abnormal development of the embryo, with increased malformation and mortality. As-SUMO-1 is a crucial regulation and modification protein resumption of embryonic diapause and early embryo development of A. sinica.
Conflict of interest statement
Figures












Similar articles
-
The Potential Roles of the Apoptosis-Related Protein PDRG1 in Diapause Embryo Restarting of Artemia sinica.Int J Mol Sci. 2018 Jan 2;19(1):126. doi: 10.3390/ijms19010126. Int J Mol Sci. 2018. PMID: 29301330 Free PMC article.
-
Cloning, expression pattern, and potential role of apoptosis inhibitor 5 in the termination of embryonic diapause and early embryo development of Artemia sinica.Gene. 2017 Sep 10;628:170-179. doi: 10.1016/j.gene.2017.07.021. Epub 2017 Jul 12. Gene. 2017. PMID: 28710039
-
Identification of the glycerol kinase gene and its role in diapause embryo restart and early embryo development of Artemia sinica.Gene. 2014 Mar 1;537(1):51-62. doi: 10.1016/j.gene.2013.12.029. Epub 2013 Dec 21. Gene. 2014. PMID: 24365596
-
Molecular chaperones, stress resistance and development in Artemia franciscana.Semin Cell Dev Biol. 2003 Oct;14(5):251-8. doi: 10.1016/j.semcdb.2003.09.019. Semin Cell Dev Biol. 2003. PMID: 14986854 Review.
-
SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems.Methods Mol Biol. 2009;497:303-17. doi: 10.1007/978-1-59745-566-4_20. Methods Mol Biol. 2009. PMID: 19107426 Review.
Cited by
-
The Potential Roles of the G1LEA and G3LEA Proteins in Early Embryo Development and in Response to Low Temperature and High Salinity in Artemia sinica.PLoS One. 2016 Sep 7;11(9):e0162272. doi: 10.1371/journal.pone.0162272. eCollection 2016. PLoS One. 2016. PMID: 27603306 Free PMC article.
-
Post-phagocytosis activation of NLRP3 inflammasome by two novel T6SS effectors.Elife. 2022 Sep 26;11:e82766. doi: 10.7554/eLife.82766. Elife. 2022. PMID: 36155655 Free PMC article.
-
The Potential Roles of the Apoptosis-Related Protein PDRG1 in Diapause Embryo Restarting of Artemia sinica.Int J Mol Sci. 2018 Jan 2;19(1):126. doi: 10.3390/ijms19010126. Int J Mol Sci. 2018. PMID: 29301330 Free PMC article.
-
RNAi-Based Therapy: Combating Shrimp Viral Diseases.Viruses. 2023 Oct 5;15(10):2050. doi: 10.3390/v15102050. Viruses. 2023. PMID: 37896827 Free PMC article. Review.
References
-
- Jiang LJ, Hou L, Zou HL, Zhang XY, Wang RF, et al. (2007) Cloning and expression analysis of p26 gene in Artemia sinica. Acta Biochem Biophys Sin 39: 351–358. - PubMed
-
- Conte FP (2007) Structure and function of the crustacean larval salt gland. Int Rev Cytol 91: 46–106.
-
- Bayer P, Arndt A, Metzger S, Mahajan R, et al. (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol 280: 275–286. - PubMed
-
- Rohit M, Christian D, Tinglu G, et al. (2007) A Small Ubiquitin-Related Polypeptide Involved in Targeting RanGAP1 to Nuclear Pore Complex Protein RanBP2. Cell 88: 98–107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

