Different walls for rods and balls: the diversity of peptidoglycan
- PMID: 24405365
- PMCID: PMC4015370
- DOI: 10.1111/mmi.12513
Different walls for rods and balls: the diversity of peptidoglycan
Abstract
Peptidoglycan performs the essential role of resisting turgor in the cell walls of most bacteria. It determines cell shape, and its biosynthesis is the target for many important antibiotics. The fundamental chemical building blocks of peptidoglycan are conserved: repeating disaccharides cross-linked by peptides. However, these blocks come in many varieties and can be assembled in different ways. So beyond the fundamental similarity, prodigious chemical, organizational and architectural diversity is revealed. Here, we track the evolution of our current understanding of peptidoglycan and underpinning technical and methodological developments. The origin and function of chemical diversity is discussed with respect to some well-studied example species. We then explore how this chemistry is manifested in elegant and complex peptidoglycan organization and how this is interpreted in different and sometimes controversial architectural models. We contend that emerging technology brings about the possibility of achieving a complete understanding of peptidoglycan chemistry, through architecture, to the way in which diverse species and populations of cells meet the challenges of maintaining viability and growth within their environmental niches, by exploiting the bioengineering versatility of peptidoglycan.
© 2014 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures

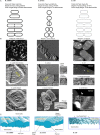
B. subtilis, sacculi broken by sonication, teichoic acids extracted, 37 000× magnification (Verwer and Nanninga, 1976).
E. coli partially digested with an endopeptidase breaking inter-peptide bonds (Verwer et al., 1978).
E. coli broken by sonication (Verwer et al., 1980).
E. coli partially digested with Cellosyl breaking intra-glycan bonds (de Pedro et al., 1997). A partial digestion with lysozyme (also breaks intra-glycan bonds) had previously been reported to leave no oriented features.

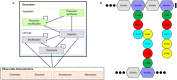
Metrics of peptidoglycan for comparison. Ranges are lowest and highest values identified in the literature (Vollmer and Seligman, ; Wheeler et al., 2011). In S. aureus and E. coli these are average values, in B. subtilis they are a representative of the overall range.
AFM gallery of sacculi comprising images comprising multiple sacculi per field, and key architectural details specific to each species (Hayhurst et al., ; Turner et al., ; 2013).
Interpretive diagrams drawn from yellow rectangles marked in ‘B’.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

