Discovery of highly potent p53-MDM2 antagonists and structural basis for anti-acute myeloid leukemia activities
- PMID: 24405416
- PMCID: PMC3985958
- DOI: 10.1021/cb400728e
Discovery of highly potent p53-MDM2 antagonists and structural basis for anti-acute myeloid leukemia activities
Abstract
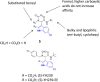
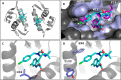
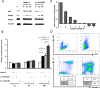
The inhibition of p53-MDM2 interaction is a promising new approach to non-genotoxic cancer treatment. A potential application for drugs blocking the p53-MDM2 interaction is acute myeloid leukemia (AML) due to the occurrence of wild type p53 (wt p53) in the majority of patients. Although there are very promising preclinical results of several p53-MDM2 antagonists in early development, none of the compounds have yet proven the utility as a next generation anticancer agent. Herein we report the design, synthesis and optimization of YH239-EE (ethyl ester of the free carboxylic acid compound YH239), a potent p53-MDM2 antagonizing and apoptosis-inducing agent characterized by a number of leukemia cell lines as well as patient-derived AML blast samples. The structural basis of the interaction between MDM2 (the p53 receptor) and YH239 is elucidated by a co-crystal structure. YH239-EE acts as a prodrug and is the most potent compound that induces apoptosis in AML cells and patient samples. The observed superior activity compared to reference compounds provides the preclinical basis for further investigation and progression of YH239-EE.
Figures


References
-
- Wang S., Zhao Y., Bernard D., Aguilar A., and Kumar S. (2009) Targeting the MDM2-p53 protein-protein interaction for new cancer therapeutics, in Protein-Protein Interactions (Wendt M. D., Ed.) Vol. 8, pp 57–79, Springer, Berlin, Heidelberg.
-
- Bueso-Ramos C. E.; Yang Y.; deLeon E.; McCown P.; Stass S. A.; Albitar M. (1993) The human MDM-2 oncogene is overexpressed in leukemias. Blood 82, 2617–2623. - PubMed
-
- Joerger A. C.; Fersht A. R. (2008) Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

