HpaP modulates type III effector secretion in Ralstonia solanacearum and harbours a substrate specificity switch domain essential for virulence
- PMID: 24405562
- PMCID: PMC6638691
- DOI: 10.1111/mpp.12119
HpaP modulates type III effector secretion in Ralstonia solanacearum and harbours a substrate specificity switch domain essential for virulence
Abstract
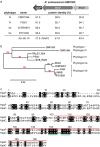
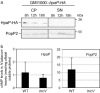
Many pathogenic bacteria have evolved a type III secretion system (T3SS) to successfully invade their host. This extracellular apparatus allows the translocation of proteins, called type III effectors (T3Es), directly into the host cells. T3Es are virulence factors that have been shown to interfere with the host's immunity or to provide nutrients from the host to the bacteria. The Gram-negative bacterium Ralstonia solanacearum is a worldwide major crop pest whose virulence strongly relies on the T3SS. In R. solanacearum, transcriptional regulation has been extensively studied. However, very few data are available concerning the role played by type III-associated regulators, such as type III chaperones and T3SS control proteins. Here, we characterized HpaP, a putative type III secretion substrate specificity switch (T3S4) protein of R. solanacearum which is not secreted by the bacterium or translocated in the plant cells. HpaP self-interacts and interacts with the PopP1 T3E. HpaP modulates the secretion of early (HrpY pilin) and late (AvrA and PopP1 T3Es) type III substrates. HpaP is dispensable for the translocation of T3Es into the host cells. Finally, we identified two regions of five amino acids in the T3S4 domain that are essential for efficient PopP1 secretion and for HpaP's role in virulence on tomato and Arabidopsis thaliana, but not required for HpaP-HpaP and HpaP-PopP1 interactions. Taken together, our results indicate that HpaP is a putative R. solanacearum T3S4 protein important for full pathogenicity on several hosts, acting as a helper for PopP1 secretion, and repressing AvrA and HrpY secretion.
Keywords: T3Es (type III effectors); T3S4 (type III secretion substrate specificity switch); T3SS (type III secretion system).; bacterial wilt; pathogenicity; secretion; translocation.
© 2014 BSPP AND JOHN WILEY & SONS LTD.
Figures

Similar articles
-
Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors.Int J Mol Sci. 2025 Apr 13;26(8):3686. doi: 10.3390/ijms26083686. Int J Mol Sci. 2025. PMID: 40332227 Free PMC article. Review.
-
HpaP Sequesters HrpJ, an Essential Component of Ralstonia solanacearum Virulence That Triggers Necrosis in Arabidopsis.Mol Plant Microbe Interact. 2020 Feb;33(2):200-211. doi: 10.1094/MPMI-05-19-0139-R. Epub 2019 Dec 16. Mol Plant Microbe Interact. 2020. PMID: 31567040
-
Study of natural diversity in response to a key pathogenicity regulator of Ralstonia solanacearum reveals new susceptibility genes in Arabidopsis thaliana.Mol Plant Pathol. 2022 Mar;23(3):321-338. doi: 10.1111/mpp.13135. Epub 2021 Dec 23. Mol Plant Pathol. 2022. PMID: 34939305 Free PMC article.
-
Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants.Proc Natl Acad Sci U S A. 2006 Sep 26;103(39):14620-5. doi: 10.1073/pnas.0509393103. Epub 2006 Sep 18. Proc Natl Acad Sci U S A. 2006. PMID: 16983093 Free PMC article.
-
The large, diverse, and robust arsenal of Ralstonia solanacearum type III effectors and their in planta functions.Mol Plant Pathol. 2020 Oct;21(10):1377-1388. doi: 10.1111/mpp.12977. Epub 2020 Aug 8. Mol Plant Pathol. 2020. PMID: 32770627 Free PMC article. Review.
Cited by
-
Comparative Secretome Analysis of Ralstonia solanacearum Type 3 Secretion-Associated Mutants Reveals a Fine Control of Effector Delivery, Essential for Bacterial Pathogenicity.Mol Cell Proteomics. 2016 Feb;15(2):598-613. doi: 10.1074/mcp.M115.051078. Epub 2015 Dec 3. Mol Cell Proteomics. 2016. PMID: 26637540 Free PMC article.
-
An atypical NLR gene confers bacterial wilt susceptibility in Arabidopsis.Plant Commun. 2023 Sep 11;4(5):100607. doi: 10.1016/j.xplc.2023.100607. Epub 2023 Apr 25. Plant Commun. 2023. PMID: 37098653 Free PMC article.
-
Quantitative Disease Resistance under Elevated Temperature: Genetic Basis of New Resistance Mechanisms to Ralstonia solanacearum.Front Plant Sci. 2017 Aug 22;8:1387. doi: 10.3389/fpls.2017.01387. eCollection 2017. Front Plant Sci. 2017. PMID: 28878784 Free PMC article.
-
Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors.Int J Mol Sci. 2025 Apr 13;26(8):3686. doi: 10.3390/ijms26083686. Int J Mol Sci. 2025. PMID: 40332227 Free PMC article. Review.
-
Pseudomonas syringae Type III Secretion Protein HrpP Manipulates Plant Immunity To Promote Infection.Microbiol Spectr. 2023 Jun 15;11(3):e0514822. doi: 10.1128/spectrum.05148-22. Epub 2023 Apr 17. Microbiol Spectr. 2023. PMID: 37067445 Free PMC article.
References
-
- Agrain, C. , Callebaut, I. , Journet, L. , Sorg, I. , Paroz, C. , Mota, L.J. and Cornelis, G.R. (2005) Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica . Mol. Microbiol. 56, 54–67. - PubMed
-
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA, 103, 14 620–14 625. - PMC - PubMed
-
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidman, J.G. , Smith, J.A. and Struhl, K. (1989) Current Protocols in Molecular Biology. New York: Greene Publishing Associates & Wiley Interscience.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

