Discovery of novel inhibitors of HIV-1 reverse transcriptase through virtual screening of experimental and theoretical ensembles
- PMID: 24405985
- PMCID: PMC3999242
- DOI: 10.1111/cbdd.12277
Discovery of novel inhibitors of HIV-1 reverse transcriptase through virtual screening of experimental and theoretical ensembles
Abstract
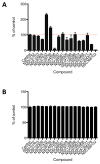
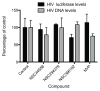
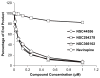
Non-nucleoside reverse transcriptase inhibitors (NNRTIs) are potent anti-HIV chemotherapeutics. Although there are FDA-approved NNRTIs, challenges such as the development of resistance have limited their utility. Here, we describe the identification of novel NNRTIs through a combination of computational and experimental approaches. Based on the known plasticity of the NNRTI binding pocket (NNIBP), we adopted an ensemble-based virtual screening strategy: coupling receptor conformations from 10 X-ray crystal structures with 120 snapshots from a total of 480 ns of molecular dynamics (MD) trajectories. A screening library of 2864 National Cancer Institute (NCI) compounds was built and docked against the ensembles in a hierarchical fashion. Sixteen diverse compounds were tested for their ability to block HIV infection in human tissue cultures using a luciferase-based reporter assay. Three promising compounds were further characterized, using a HIV-1 RT-based polymerase assay, to determine the specific mechanism of inhibition. We found that 2 of the three compounds inhibited the polymerase activity of RT (with potency similar to the positive control, the FDA-approved drug nevirapine). Through a computational approach, we were able to discover two compounds which inhibit HIV replication and block the activity of RT, thus offering the potential for optimization into mature inhibitors.
Keywords: HIV; NNRTI; molecular dynamics; reverse transcriptase; virtual screening.
© 2014 John Wiley & Sons A/S.
Figures






Similar articles
-
Application of Structure-based Methods to Analyze Resistance Mutations for Chemically Diverse Non-Nucleoside Reverse Transcriptase Inhibitors.Curr HIV Res. 2020;18(4):283-291. doi: 10.2174/1570162X18666200603141209. Curr HIV Res. 2020. PMID: 32493197
-
Elucidating the inhibition mechanism of HIV-1 non-nucleoside reverse transcriptase inhibitors through multicopy molecular dynamics simulations.J Mol Biol. 2009 May 8;388(3):644-58. doi: 10.1016/j.jmb.2009.03.037. Epub 2009 Mar 24. J Mol Biol. 2009. PMID: 19324058 Free PMC article.
-
Novel natural non-nucleoside inhibitors of HIV-1 reverse transcriptase identified by shape- and structure-based virtual screening techniques.Eur J Med Chem. 2019 Jan 1;161:1-10. doi: 10.1016/j.ejmech.2018.10.029. Epub 2018 Oct 13. Eur J Med Chem. 2019. PMID: 30342421
-
Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors.Virus Res. 2008 Jun;134(1-2):147-56. doi: 10.1016/j.virusres.2008.01.002. Epub 2008 Mar 26. Virus Res. 2008. PMID: 18372072 Free PMC article. Review.
-
Current status of the non-nucleoside reverse transcriptase inhibitors of human immunodeficiency virus type 1.Curr Top Med Chem. 2004;4(9):921-44. doi: 10.2174/1568026043388420. Curr Top Med Chem. 2004. PMID: 15134549 Review.
Cited by
-
Stimuli-sensitive thiolated hyaluronic acid based nanofibers: synthesis, preclinical safety and in vitro anti-HIV activity.Nanomedicine (Lond). 2016 Nov;11(22):2935-2958. doi: 10.2217/nnm-2016-0103. Epub 2016 Oct 27. Nanomedicine (Lond). 2016. PMID: 27785967 Free PMC article.
-
Enrichment of chemical libraries docked to protein conformational ensembles and application to aldehyde dehydrogenase 2.J Chem Inf Model. 2014 Jul 28;54(7):2105-16. doi: 10.1021/ci5002026. Epub 2014 Jun 30. J Chem Inf Model. 2014. PMID: 24856086 Free PMC article.
-
Molecular docking based design of Inhibitors for viral Non-Nucleosidase as potential anti-retroviral agents.Bioinformation. 2020 Oct 31;16(10):736-741. doi: 10.6026/97320630016736. eCollection 2020. Bioinformation. 2020. PMID: 34675458 Free PMC article.
-
Computational drug design strategies applied to the modelling of human immunodeficiency virus-1 reverse transcriptase inhibitors.Mem Inst Oswaldo Cruz. 2015 Nov;110(7):847-64. doi: 10.1590/0074-02760150239. Mem Inst Oswaldo Cruz. 2015. PMID: 26560977 Free PMC article. Review.
-
Enhancing Virtual Screening Performance of Protein Kinases with Molecular Dynamics Simulations.J Chem Inf Model. 2016 Oct 24;56(10):1923-1935. doi: 10.1021/acs.jcim.6b00261. Epub 2016 Oct 3. J Chem Inf Model. 2016. PMID: 27662181 Free PMC article.
References
-
- Das K, Arnold E, Hughes S. Nonnucleoside Reverse Transcriptase Inhibitors (NNRTIs) In: LeGrice S, Gotte M, editors. Human Immunodeficiency Virus Reverse Transcriptase. Springer; New York: 2013. pp. 123–39.
-
- Zhan P, Chen X, Li D, Fang Z, De Clercq E, Liu X. HIV-1 NNRTIs: structural diversity, pharmacophore similarity, and implications for drug design. Med Res Rev. 2013 Jun;33( Suppl 1):E1–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

