Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity
- PMID: 24406253
- PMCID: PMC3954709
- DOI: 10.1016/j.mad.2013.12.008
Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity
Abstract
Cockayne Syndrome is a segmental premature aging syndrome, which can be caused by loss of function of the CSB protein. CSB is essential for genome maintenance and has numerous interaction partners with established roles in different DNA repair pathways including transcription coupled nucleotide excision repair and base excision repair. Here, we describe a new interaction partner for CSB, the DNA glycosylase NEIL2. Using both cell extracts and recombinant proteins, CSB and NEIL2 were found to physically interact independently of DNA. We further found that CSB is able to stimulate NEIL2 glycosylase activity on a 5-hydroxyl uracil lesion in a DNA bubble structure substrate in vitro. A novel 4,6-diamino-5-formamidopyrimidine (FapyA) specific incision activity of NEIL2 was also stimulated by CSB. To further elucidate the biological role of the interaction, immunofluorescence studies were performed, showing an increase in cytoplasmic CSB and NEIL2 co-localization after oxidative stress. Additionally, stalling of the progression of the transcription bubble with α-amanitin resulted in increased co-localization of CSB and NEIL2. Finally, CSB knockdown resulted in reduced incision of 8-hydroxyguanine in a DNA bubble structure using whole cell extracts. Taken together, our data supports a biological role for CSB and NEIL2 in transcription associated base excision repair.
Keywords: Base excision repair; CSB; Cockayne Syndrome; NEIL2; Oxidative damage.
Copyright © 2014 Elsevier Ireland Ltd. All rights reserved.
Figures






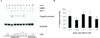


References
-
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, 3rd, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

