Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory
- PMID: 24406459
- PMCID: PMC4079763
- DOI: 10.1038/gim.2013.183
Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory
Abstract
Purpose: Sanger sequencing is currently considered the gold standard methodology for clinical molecular diagnostic testing. However, next-generation sequencing has already emerged as a much more efficient means to identify genetic variants within gene panels, the exome, or the genome. We sought to assess the accuracy of next-generation sequencing variant identification in our clinical genomics laboratory with the goal of establishing a quality score threshold for confirmatory Sanger-based testing.
Methods: Confirmation data for reported results from 144 sequential clinical exome-sequencing cases (94 unique variants) and an additional set of 16 variants from comparable research samples were analyzed.
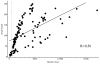
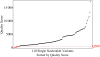
Results: Of the 110 total single-nucleotide variants analyzed, 103 variants had a quality score ≥Q500, 103 (100%) of which were confirmed by Sanger sequencing. Of the remaining seven variants with quality scores <Q500, six were confirmed by Sanger sequencing (85%).
Conclusion: For single-nucleotide variants, we predict that going forward, we will be able to reduce our Sanger confirmation workload by 70-80%. This serves as a proof of principle that as long as sufficient validation and quality control measures are implemented, the volume of Sanger confirmation can be reduced, alleviating a significant amount of the labor and cost burden on clinical laboratories wishing to use next-generation sequencing technology. However, Sanger confirmation of low-quality single-nucleotide variants and all insertions or deletions <10 bp remains necessary at this time in our laboratory.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

