Gastrointestinal malignancy and the microbiome
- PMID: 24406471
- PMCID: PMC3995897
- DOI: 10.1053/j.gastro.2014.01.001
Gastrointestinal malignancy and the microbiome
Abstract
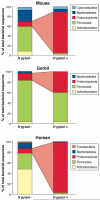
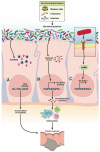
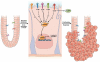
Microbial species participate in the genesis of a substantial number of malignancies-in conservative estimates, at least 15% of all cancer cases are attributable to infectious agents. Little is known about the contribution of the gastrointestinal microbiome to the development of malignancies. Resident microbes can promote carcinogenesis by inducing inflammation, increasing cell proliferation, altering stem cell dynamics, and producing metabolites such as butyrate, which affect DNA integrity and immune regulation. Studies in human beings and rodent models of cancer have identified effector species and relationships among members of the microbial community in the stomach and colon that increase the risk for malignancy. Strategies to manipulate the microbiome, or the immune response to such bacteria, could be developed to prevent or treat certain gastrointestinal cancers.
Keywords: Bacteria; Cancer; Inflammation.
Copyright © 2014 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncology. 2012;13:607–15. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

