A novel unsaturated β-glucuronyl hydrolase involved in ulvan degradation unveils the versatility of stereochemistry requirements in family GH105
- PMID: 24407291
- PMCID: PMC3937685
- DOI: 10.1074/jbc.M113.537480
A novel unsaturated β-glucuronyl hydrolase involved in ulvan degradation unveils the versatility of stereochemistry requirements in family GH105
Abstract
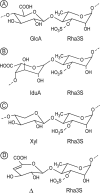
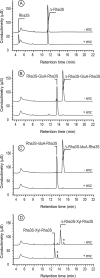
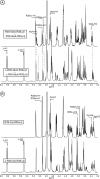
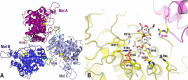
Ulvans are cell wall matrix polysaccharides in green algae belonging to the genus Ulva. Enzymatic degradation of the polysaccharide by ulvan lyases leads to the production of oligosaccharides with an unsaturated β-glucuronyl residue located at the non-reducing end. Exploration of the genomic environment around the Nonlabens ulvanivorans (previously Percicivirga ulvanivorans) ulvan lyase revealed a gene highly similar to known unsaturated uronyl hydrolases classified in the CAZy glycoside hydrolase family 105. The gene was cloned, the protein was overexpressed in Escherichia coli, and enzymology experiments demonstrated its unsaturated β-glucuronyl activity. Kinetic analysis of purified oligo-ulvans incubated with the new enzyme showed that the full substrate specificity is attained by three subsites that preferentially bind anionic residues (sulfated rhamnose, glucuronic/iduronic acid). The three-dimensional crystal structure of the native enzyme reveals that a trimeric organization is required for substrate binding and recognition at the +2 binding subsite. This novel unsaturated β-glucuronyl hydrolase is part of a previously uncharacterized subgroup of GH105 members and exhibits only a very limited sequence similarity to known unsaturated β-glucuronyl sequences previously found only in family GH88. Clan-O formed by families GH88 and GH105 was singular in the fact that it covered families acting on both axial and equatorial glycosidic linkages, respectively. The overall comparison of active site structures between enzymes from these two families highlights how that within family GH105, and unlike for classical glycoside hydrolysis, the hydrolysis of vinyl ether groups from unsaturated saccharides occurs independently of the α or β configuration of the cleaved linkage.
Keywords: Biodegradation; Crystal Structure; Enzyme Catalysis; GH105; Glycoside Hydrolases; Polysaccharide; Ulvan.
Figures




References
-
- Lahaye M., Robic A. (2007) Structure and functional properties of ulvan, a polysaccharide from green seaweeds. Biomacromolecules 8, 1765–1774 - PubMed
-
- Percival E., McDowell R. H. (1967) Chemistry and enzymology of marine algal polysaccharides, Academic Press Inc., London
-
- Quemener B., Lahaye M., Bobin-Dubigeon C. (1997) Sugar determination in ulvans by a chemical-enzymatic method coupled to high performance anion exchange chromatography. J. Appl. Phycol. 9, 179–188
-
- Ray B., Lahaye M. (1995) Cell wall polysaccharides from the marine green alga Ulva “rigida” (Ulvales, Chlorophyta). Extraction and chemical composition. Carbohydr. Res. 274, 313–318 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

