Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase
- PMID: 24407421
- PMCID: PMC3915904
- DOI: 10.1038/cr.2014.3
Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase
Abstract
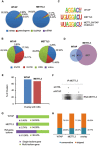
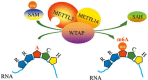
The methyltransferase like 3 (METTL3)-containing methyltransferase complex catalyzes the N6-methyladenosine (m6A) formation, a novel epitranscriptomic marker; however, the nature of this complex remains largely unknown. Here we report two new components of the human m6A methyltransferase complex, Wilms' tumor 1-associating protein (WTAP) and methyltransferase like 14 (METTL14). WTAP interacts with METTL3 and METTL14, and is required for their localization into nuclear speckles enriched with pre-mRNA processing factors and for catalytic activity of the m6A methyltransferase in vivo. The majority of RNAs bound by WTAP and METTL3 in vivo represent mRNAs containing the consensus m6A motif. In the absence of WTAP, the RNA-binding capability of METTL3 is strongly reduced, suggesting that WTAP may function to regulate recruitment of the m6A methyltransferase complex to mRNA targets. Furthermore, transcriptomic analyses in combination with photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) illustrate that WTAP and METTL3 regulate expression and alternative splicing of genes involved in transcription and RNA processing. Morpholino-mediated knockdown targeting WTAP and/or METTL3 in zebrafish embryos caused tissue differentiation defects and increased apoptosis. These findings provide strong evidence that WTAP may function as a regulatory subunit in the m6A methyltransferase complex and play a critical role in epitranscriptomic regulation of RNA metabolism.
Figures



References
-
- Chen-Kiang S, Nevins JR, Darnell JE., Jr N-6-methyl-adenosine in adenovirus type 2 nuclear RNA is conserved in the formation of messenger RNA. J Mol Biol. 1979;135:733–752. - PubMed
-
- Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus. Biochemistry. 1975;14:4367–4374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

