Motif-directed redesign of enzyme specificity
- PMID: 24407908
- PMCID: PMC3945839
- DOI: 10.1002/pro.2417
Motif-directed redesign of enzyme specificity
Abstract
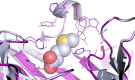
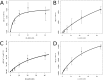
Computational protein design relies on several approximations, including the use of fixed backbones and rotamers, to reduce protein design to a computationally tractable problem. However, allowing backbone and off-rotamer flexibility leads to more accurate designs and greater conformational diversity. Exhaustive sampling of this additional conformational space is challenging, and often impossible. Here, we report a computational method that utilizes a preselected library of native interactions to direct backbone flexibility to accommodate placement of these functional contacts. Using these native interaction modules, termed motifs, improves the likelihood that the interaction can be realized, provided that suitable backbone perturbations can be identified. Furthermore, it allows a directed search of the conformational space, reducing the sampling needed to find low energy conformations. We implemented the motif-based design algorithm in Rosetta, and tested the efficacy of this method by redesigning the substrate specificity of methionine aminopeptidase. In summary, native enzymes have evolved to catalyze a wide range of chemical reactions with extraordinary specificity. Computational enzyme design seeks to generate novel chemical activities by altering the target substrates of these existing enzymes. We have implemented a novel approach to redesign the specificity of an enzyme and demonstrated its effectiveness on a model system.
Keywords: computational protein design; enzyme design; molecular specificity; mutagenesis.
© 2014 The Authors Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Figures




References
-
- Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. - PubMed
-
- Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. - PubMed
-
- Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. - PubMed
-
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

