Green-lighting green fluorescent protein: faster and more efficient folding by eliminating a cis-trans peptide isomerization event
- PMID: 24408076
- PMCID: PMC3970891
- DOI: 10.1002/pro.2421
Green-lighting green fluorescent protein: faster and more efficient folding by eliminating a cis-trans peptide isomerization event
Abstract
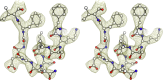
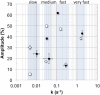
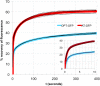
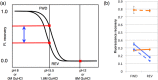
Wild-type green fluorescent protein (GFP) folds on a time scale of minutes. The slow step in folding is a cis-trans peptide bond isomerization. The only conserved cis-peptide bond in the native GFP structure, at P89, was remodeled by the insertion of two residues, followed by iterative energy minimization and side chain design. The engineered GFP was synthesized and found to fold faster and more efficiently than its template protein, recovering 50% more of its fluorescence upon refolding. The slow phase of folding is faster and smaller in amplitude, and hysteresis in refolding has been eliminated. The elimination of a previously reported kinetically trapped state in refolding suggests that X-P89 is trans in the trapped state. A 2.55 Å resolution crystal structure revealed that the new variant contains only trans-peptide bonds, as designed. This is the first instance of a computationally remodeled fluorescent protein that folds faster and more efficiently than wild type.
Keywords: GFP; cis; folding kinetics; protein design; trans isomerization.
© 2014 The Protein Society.
Figures

References
-
- Tsien RY. The green fluorescent protein. Ann Rev Biochem. 1998;67:509–544. - PubMed
-
- Zimmer M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem Rev. 2002;102:759–781. - PubMed
-
- Crone DE, Huang YM, Pitman DJ, Schenkelberg C, Fraser K, Macari S, Bystroff C. GFP-Based biosensors. In: Rinken T, editor. State of the art in biosensors—General aspects. InTech; 2013. ISBN: 978-953-51-1004-0.
-
- Zimmer M. GFP: From jellyfish to the Nobel prize and beyond. Chem Soc Rev. 2009;38:2823. - PubMed
-
- Jackson SE, Craggs TD, Huang J. Understanding the folding of GFP using biophysical techniques. Expert Rev Proteom. 2006;3:545–559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

