Comparative genome analysis of the closely related Synechocystis strains PCC 6714 and PCC 6803
- PMID: 24408876
- PMCID: PMC4060947
- DOI: 10.1093/dnares/dst055
Comparative genome analysis of the closely related Synechocystis strains PCC 6714 and PCC 6803
Abstract
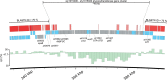
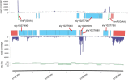
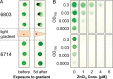
Synechocystis sp. PCC 6803 is the most popular cyanobacterial model for prokaryotic photosynthesis and for metabolic engineering to produce biofuels. Genomic and transcriptomic comparisons between closely related bacteria are powerful approaches to infer insights into their metabolic potentials and regulatory networks. To enable a comparative approach, we generated the draft genome sequence of Synechocystis sp. PCC 6714, a closely related strain of 6803 (16S rDNA identity 99.4%) that also is amenable to genetic manipulation. Both strains share 2838 protein-coding genes, leaving 845 unique genes in Synechocystis sp. PCC 6803 and 895 genes in Synechocystis sp. PCC 6714. The genetic differences include a prophage in the genome of strain 6714, a different composition of the pool of transposable elements, and a ∼ 40 kb genomic island encoding several glycosyltransferases and transport proteins. We verified several physiological differences that were predicted on the basis of the respective genome sequence. Strain 6714 exhibited a lower tolerance to Zn(2+) ions, associated with the lack of a corresponding export system and a lowered potential of salt acclimation due to the absence of a transport system for the re-uptake of the compatible solute glucosylglycerol. These new data will support the detailed comparative analyses of this important cyanobacterial group than has been possible thus far. Genome information for Synechocystis sp. PCC 6714 has been deposited in Genbank (accession no AMZV01000000).
Keywords: comparative genomics; cyanophages; genome sequence; prophage; salt acclimation.
© The Author 2014. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures

Similar articles
-
Genomic structure of the cyanobacterium Synechocystis sp. PCC 6803 strain GT-S.DNA Res. 2011 Oct;18(5):393-9. doi: 10.1093/dnares/dsr026. Epub 2011 Jul 29. DNA Res. 2011. PMID: 21803841 Free PMC article.
-
Coexpression of Tail Fiber and Tail Protein Genes of the Cyanophage PP Using a Synthetic Genomics Approach Enhances the Salt Tolerance of Synechocystis PCC 6803.Microbiol Spectr. 2023 Jun 15;11(3):e0500922. doi: 10.1128/spectrum.05009-22. Epub 2023 May 1. Microbiol Spectr. 2023. PMID: 37125914 Free PMC article.
-
Diurnal Regulation of Cellular Processes in the Cyanobacterium Synechocystis sp. Strain PCC 6803: Insights from Transcriptomic, Fluxomic, and Physiological Analyses.mBio. 2016 May 3;7(3):e00464-16. doi: 10.1128/mBio.00464-16. mBio. 2016. PMID: 27143387 Free PMC article.
-
Development of Synechocystis sp. PCC 6803 as a phototrophic cell factory.Mar Drugs. 2013 Aug 13;11(8):2894-916. doi: 10.3390/md11082894. Mar Drugs. 2013. PMID: 23945601 Free PMC article. Review.
-
Current knowledge and recent advances in understanding metabolism of the model cyanobacterium Synechocystis sp. PCC 6803.Biosci Rep. 2020 Apr 30;40(4):BSR20193325. doi: 10.1042/BSR20193325. Biosci Rep. 2020. PMID: 32149336 Free PMC article. Review.
Cited by
-
Salt acclimation of cyanobacteria and their application in biotechnology.Life (Basel). 2014 Dec 29;5(1):25-49. doi: 10.3390/life5010025. Life (Basel). 2014. PMID: 25551682 Free PMC article. Review.
-
Insights into the Planktothrix genus: Genomic and metabolic comparison of benthic and planktic strains.Sci Rep. 2017 Jan 24;7:41181. doi: 10.1038/srep41181. Sci Rep. 2017. PMID: 28117406 Free PMC article.
-
Modes of Fatty Acid desaturation in cyanobacteria: an update.Life (Basel). 2015 Feb 16;5(1):554-67. doi: 10.3390/life5010554. Life (Basel). 2015. PMID: 25809965 Free PMC article.
-
The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7.Proc Natl Acad Sci U S A. 2015 Nov 10;112(45):E6243-52. doi: 10.1073/pnas.1508412112. Epub 2015 Oct 22. Proc Natl Acad Sci U S A. 2015. PMID: 26494284 Free PMC article.
-
Molar-based targeted metabolic profiling of cyanobacterial strains with potential for biological production.Metabolites. 2014 Jun 20;4(2):499-516. doi: 10.3390/metabo4020499. Metabolites. 2014. PMID: 24957038 Free PMC article.
References
-
- Kaneko T., Sato S., Kotani H., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement) DNA Res. 1996;3:185–209. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

