Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories
- PMID: 24408911
- PMCID: PMC3969563
- DOI: 10.1093/molbev/msu040
Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories
Abstract
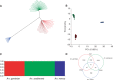
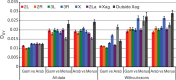
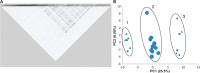
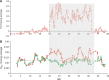
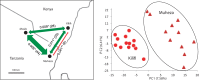
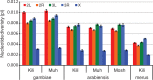
Anopheles gambiae s.l. are important malaria vectors, but little is known about their genomic variation in the wild. Here, we present inter- and intraspecies analysis of genome-wide RADseq data, in three Anopheles gambiae s.l. species collected from East Africa. The mosquitoes fall into three genotypic clusters representing described species (A. gambiae, A. arabiensis, and A. merus) with no evidence of cryptic breeding units. Anopheles merus is the most divergent of the three species, supporting a recent new phylogeny based on chromosomal inversions. Even though the species clusters are well separated, there is extensive shared polymorphism, particularly between A. gambiae and A. arabiensis. Divergence between A. gambiae and A. arabiensis does not vary across the autosomes but is higher in X-linked inversions than elsewhere on X or on the autosomes, consistent with the suggestion that this inversion (or a gene within it) is important in reproductive isolation between the species. The 2La/2L+(a) inversion shows no more evidence of introgression between A. gambiae and A. arabiensis than the rest of the autosomes. Population differentiation within A. gambiae and A. arabiensis is weak over approximately 190-270 km, implying no strong barriers to dispersal. Analysis of Tajima's D and the allele frequency spectrum is consistent with modest population increases in A. arabiensis and A. merus, but a more complex demographic history of expansion followed by contraction in A. gambiae. Although they are less than 200 km apart, the two A. gambiae populations show evidence of different demographic histories.
Keywords: 2La; Anopheles arabiensis; Anopheles gambiae; Anopheles merus; RADseq; population genomics.
Figures

References
-
- Andolfatto P, Depaulis F, Navarro A. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res. 2001;77:1–8. - PubMed
-
- Arnold B, Corbett-Detig RB, Hartl D, Bomblies K. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol. 2013;22:3179–3190. - PubMed
-
- Bachtrog D. Evidence for male-driven evolution in Drosophila. Mol Biol Evol. 2008;25:617–619. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

