Probing the subcellular localization of hopanoid lipids in bacteria using NanoSIMS
- PMID: 24409299
- PMCID: PMC3883690
- DOI: 10.1371/journal.pone.0084455
Probing the subcellular localization of hopanoid lipids in bacteria using NanoSIMS
Abstract
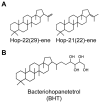
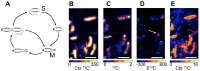
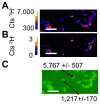
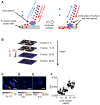
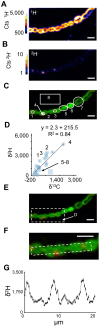
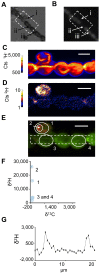
The organization of lipids within biological membranes is poorly understood. Some studies have suggested lipids group into microdomains within cells, but the evidence remains controversial due to non-native imaging techniques. A recently developed NanoSIMS technique indicated that sphingolipids group into microdomains within membranes of human fibroblast cells. We extended this NanoSIMS approach to study the localization of hopanoid lipids in bacterial cells by developing a stable isotope labeling method to directly detect subcellular localization of specific lipids in bacteria with ca. 60 nm resolution. Because of the relatively small size of bacterial cells and the relative abundance of hopanoid lipids in membranes, we employed a primary (2)H-label to maximize our limit of detection. This approach permitted the analysis of multiple stable isotope labels within the same sample, enabling visualization of subcellular lipid microdomains within different cell types using a secondary label to mark the growing end of the cell. Using this technique, we demonstrate subcellular localization of hopanoid lipids within alpha-proteobacterial and cyanobacterial cells. Further, we provide evidence of hopanoid lipid domains in between cells of the filamentous cyanobacterium Nostoc punctiforme. More broadly, our method provides a means to image lipid microdomains in a wide range of cell types and test hypotheses for their functions in membranes.
Conflict of interest statement
Figures
References
-
- Owens DM, Magenau A, Williamson D & Gaus K (2012) The lipid raft hypothesis revisited–new insights on raft composition and function from super-resolution fluorescence microscopy. Bioessays 34: 739–747. - PubMed
-
- Suzuki KG (2004) Lipid rafts generate digital-like signal transduction in cell plasma membranes. Biotechnol J 7: 753–761. - PubMed
-
- Chen JC, Viollier PH, Shapiro L (2004) A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Micro 55: 1085–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

