Application of a high-resolution mass-spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures
- PMID: 24410521
- PMCID: PMC3982966
- DOI: 10.1021/ac403565m
Application of a high-resolution mass-spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures
Abstract
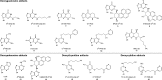
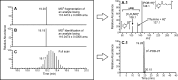
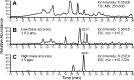
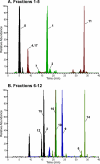
Liquid chromatography coupled with mass spectrometry (LC-MS) is the method of choice for analysis of covalent modification of DNA. DNA adductomics is an extension of this approach allowing for the screening for both known and unknown DNA adducts. In the research reported here, a new high-resolution/accurate mass MS(n) methodology has been developed representing an important advance for the investigation of in vivo biological samples and for the assessment of DNA damage from various human exposures. The methodology was tested and optimized using a mixture of 18 DNA adducts representing a range of biologically relevant modifications on all four bases and using DNA from liver tissue of mice exposed to the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). In the latter experiment, previously characterized adducts, both expected and unexpected, were observed.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

