Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy
- PMID: 24410571
- PMCID: PMC4241876
- DOI: 10.1089/ars.2013.5795
Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy
Abstract
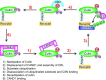
Significance: Protein neddylation is catalyzed by an E1 NEDD8-activating enzyme (NAE), an E2 NEDD8-conjugating enzyme, and an E3 NEDD8 ligase. Known physiological substrates of neddylation are cullin family members. Cullin neddylation leads to activation of cullin-RING ligases (CRLs), the largest family of E3 ubiquitin ligases responsible for ubiquitylation and degradation of many key signaling/regulatory proteins. Thus, through modulating CRLs, neddylation regulates many biological processes, including cell cycle progression, signal transduction, and tumorigenesis. Given that NEDD8 is overexpressed and CRLs are abnormally activated in many human cancers, targeting protein neddylation, in general, and cullin neddylation, in particular, appears to be an attractive anticancer approach.
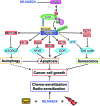
Recent advances: MLN4924, a small molecule inhibitor of NAE, was discovered that inactivates CRLs and causes accumulation of CRL substrates to suppress tumor cell growth both in vitro and in vivo. Promising preclinical results advanced MLN4924 to several clinical trials for anticancer therapy.
Critical issues: In preclinical settings, MLN4924 effectively suppresses tumor cell growth by inducing apoptosis, senescence, and autophagy, and causes sensitization to chemoradiation therapies in a cellular context-dependent manner. Signal molecules that determine the cell fate upon MLN4924 treatment, however, remain elusive. Cancer cells develop MLN4924 resistance by selecting target mutations.
Future directions: In the clinical side, several Phase 1b trials are under way to determine the safety and efficacy of MLN4924, acting alone or in combination with conventional chemotherapy, against human solid tumors. In the preclinical side, the efforts are being made to develop additional neddylation inhibitors by targeting NEDD8 E2s and E3s.
Figures




References
-
- Bech-Otschir D, Seeger M, and Dubiel W. The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci 115: 467–473, 2002 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

