Cytogenomic mapping and bioinformatic mining reveal interacting brain expressed genes for intellectual disability
- PMID: 24410907
- PMCID: PMC3905969
- DOI: 10.1186/1755-8166-7-4
Cytogenomic mapping and bioinformatic mining reveal interacting brain expressed genes for intellectual disability
Abstract
Background: Microarray analysis has been used as the first-tier genetic testing to detect chromosomal imbalances and copy number variants (CNVs) for pediatric patients with intellectual and developmental disabilities (ID/DD). To further investigate the candidate genes and underlying dosage-sensitive mechanisms related to ID, cytogenomic mapping of critical regions and bioinformatic mining of candidate brain-expressed genes (BEGs) and their functional interactions were performed. Critical regions of chromosomal imbalances and pathogenic CNVs were mapped by subtracting known benign CNVs from the Databases of Genomic Variants (DGV) and extracting smallest overlap regions with cases from DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER). BEGs from these critical regions were revealed by functional annotation using Database for Annotation, Visualization, and Integrated Discovery (DAVID) and by tissue expression pattern from Uniprot. Cross-region interrelations and functional networks of the BEGs were analyzed using Gene Relationships Across Implicated Loci (GRAIL) and Ingenuity Pathway Analysis (IPA).
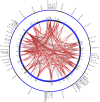
Results: Of the 1,354 patients analyzed by oligonucleotide array comparative genomic hybridization (aCGH), pathogenic abnormalities were detected in 176 patients including genomic disorders in 66 patients (37.5%), subtelomeric rearrangements in 45 patients (25.6%), interstitial imbalances in 33 patients (18.8%), chromosomal structural rearrangements in 17 patients (9.7%) and aneuploidies in 15 patients (8.5%). Subtractive and extractive mapping defined 82 disjointed critical regions from the detected abnormalities. A total of 461 BEGs was generated from 73 disjointed critical regions. Enrichment of central nervous system specific genes in these regions was noted. The number of BEGs increased with the size of the regions. A list of 108 candidate BEGs with significant cross region interrelation was identified by GRAIL and five significant gene networks involving cell cycle, cell-to-cell signaling, cellular assembly, cell morphology, and gene expression regulations were denoted by IPA.
Conclusions: These results characterized ID related cross-region interrelations and multiple networks of candidate BEGs from the detected genomic imbalances. Further experimental study of these BEGs and their interactions will lead to a better understanding of dosage-sensitive mechanisms and modifying effects of human mental development.
Figures

Similar articles
-
Spectrum of Cytogenomic Abnormalities Revealed by Array Comparative Genomic Hybridization on Products of Conception Culture Failure and Normal Karyotype Samples.J Genet Genomics. 2016 Mar 20;43(3):121-31. doi: 10.1016/j.jgg.2016.02.002. Epub 2016 Feb 13. J Genet Genomics. 2016. PMID: 27020032
-
New insights in the interpretation of array-CGH: autism spectrum disorder and positive family history for intellectual disability predict the detection of pathogenic variants.Ital J Pediatr. 2016 Apr 12;42:39. doi: 10.1186/s13052-016-0246-7. Ital J Pediatr. 2016. PMID: 27072107 Free PMC article.
-
Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features.Eur J Paediatr Neurol. 2013 Nov;17(6):589-99. doi: 10.1016/j.ejpn.2013.04.010. Epub 2013 May 24. Eur J Paediatr Neurol. 2013. PMID: 23711909
-
The discovery of microdeletion syndromes in the post-genomic era: review of the methodology and characterization of a new 1q41q42 microdeletion syndrome.Genet Med. 2007 Sep;9(9):607-16. doi: 10.1097/gim.0b013e3181484b49. Genet Med. 2007. PMID: 17873649 Review.
-
Array comparative genomic hybridization and computational genome annotation in constitutional cytogenetics: suggesting candidate genes for novel submicroscopic chromosomal imbalance syndromes.Genet Med. 2007 Sep;9(9):642-9. doi: 10.1097/gim.0b013e318145b27b. Genet Med. 2007. PMID: 17873653 Review.
Cited by
-
Changes in and Efficacies of Indications for Invasive Prenatal Diagnosis of Cytogenomic Abnormalities: 13 Years of Experience in a Single Center.Med Sci Monit. 2015 Jul 5;21:1942-8. doi: 10.12659/MSM.893870. Med Sci Monit. 2015. PMID: 26143093 Free PMC article.
-
Genotype-Phenotype Correlations for Putative Haploinsufficient Genes in Deletions of 6q26-q27: Report of Eight Patients and Review of Literature.Glob Med Genet. 2022 Mar 11;9(2):166-174. doi: 10.1055/s-0042-1743568. eCollection 2022 Jun. Glob Med Genet. 2022. PMID: 35707784 Free PMC article.
-
Intellectual disability: dendritic anomalies and emerging genetic perspectives.Acta Neuropathol. 2021 Feb;141(2):139-158. doi: 10.1007/s00401-020-02244-5. Epub 2020 Nov 23. Acta Neuropathol. 2021. PMID: 33226471 Free PMC article. Review.
-
In silico molecular cytogenetics: a bioinformatic approach to prioritization of candidate genes and copy number variations for basic and clinical genome research.Mol Cytogenet. 2014 Dec 9;7(1):98. doi: 10.1186/s13039-014-0098-z. eCollection 2014. Mol Cytogenet. 2014. PMID: 25525469 Free PMC article.
-
A Retrospective Analysis of 10-Year Data Assessed the Diagnostic Accuracy and Efficacy of Cytogenomic Abnormalities in Current Prenatal and Pediatric Settings.Front Genet. 2019 Nov 20;10:1162. doi: 10.3389/fgene.2019.01162. eCollection 2019. Front Genet. 2019. PMID: 31850057 Free PMC article.
References
-
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ. et al.Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. - DOI - PMC - PubMed
-
- Xiang B, Li A, Valentin D, Nowak NJ, Zhao H, Li P. Analytical and clinical validity of whole-genome oligonucleotide array comparative genomic hybridization for pediatric patients with mental retardation and developmental delay. Am J Med Genet A. 2008;146A:1942–1954. doi: 10.1002/ajmg.a.32411. - DOI - PubMed
-
- Hayashi S, Imoto I, Aizu Y, Okamoto N, Mizuno S, Kurosawa K, Honda S, Araki S, Mizutani S, Numabe H. et al.Clinical application of array-based comparative genomic hybridization by two-stage screening for 536 patients with mental retardation and multiple congenital anomalies. J Hum Genet. 2010;56:110–124. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

