The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis
- PMID: 24412063
- PMCID: PMC3947554
- DOI: 10.1016/j.molcel.2013.12.009
The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis
Abstract
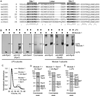
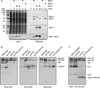
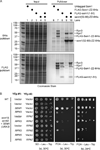
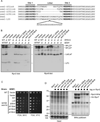
The intrinsically disordered yeast protein Sem1 (DSS1 in mammals) participates in multiple protein complexes, including the proteasome, but its role(s) within these complexes is uncertain. We report that Sem1 enforces the ordered incorporation of subunits Rpn3 and Rpn7 into the assembling proteasome lid. Sem1 uses conserved acidic segments separated by a flexible linker to grasp Rpn3 and Rpn7. The same segments are used for protein binding in other complexes, but in the proteasome lid they are uniquely deployed for recognizing separate polypeptides. We engineered TEV protease-cleavage sites into Sem1 to show that the tethering function of Sem1 is important for the biogenesis and integrity of the Rpn3-Sem1-Rpn7 ternary complex but becomes dispensable once the ternary complex incorporates into larger lid precursors. Thus, although Sem1 is a stoichiometric component of the mature proteasome, it has a distinct, chaperone-like function specific to early stages of proteasome assembly.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
-
- Bohn S, Sakata E, Beck F, Pathare GR, Schnitger J, Nagy I, Baumeister W, Forster F. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochem Biophys Res Commun. 2013;435:250–254. - PubMed
-
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature reviews. Molecular cell biology. 2005;6:197–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

