INF2-mediated severing through actin filament encirclement and disruption
- PMID: 24412206
- PMCID: PMC3992431
- DOI: 10.1016/j.cub.2013.12.018
INF2-mediated severing through actin filament encirclement and disruption
Abstract
Background: INF2 is a formin protein with the unique ability to accelerate both actin polymerization and depolymerization, the latter requiring filament severing. Mutations in INF2 lead to the kidney disease focal segmental glomerulosclerosis (FSGS) and the neurological disorder Charcot-Marie Tooth disease (CMTD).
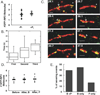
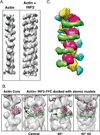
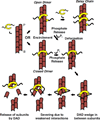
Results: Here, we compare the severing mechanism of INF2 with that of the well-studied severing protein cofilin. INF2, like cofilin, binds stoichiometrically to filament sides and severs in a manner that requires phosphate release from the filament. In contrast to cofilin, however, INF2 binds ADP and ADP-Pi filaments equally well. Furthermore, two-color total internal reflection fluorescence (TIRF) microscopy reveals that a low number of INF2 molecules, as few as a single INF2 dimer, are capable of severing, while measurable cofilin-mediated severing requires more extensive binding. Hence, INF2 is a more potent severing protein than cofilin. While a construct containing the FH1 and FH2 domains alone has some severing activity, addition of the C-terminal region increases severing potency by 40-fold, and we show that the WH2-resembling DAD motif is responsible for this increase. Helical 3D reconstruction from electron micrographs at 20 Å resolution provides a structure of filament-bound INF2, showing that the FH2 domain encircles the filament.
Conclusions: We propose a severing model in which FH2 binding and phosphate release causes local filament deformation, allowing the DAD to bind adjacent actin protomers, further disrupting filament structure.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors do not have any financial conflict of interest associated with this work.
Figures



Similar articles
-
Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing.J Biol Chem. 2012 Oct 5;287(41):34234-45. doi: 10.1074/jbc.M112.365122. Epub 2012 Aug 9. J Biol Chem. 2012. PMID: 22879592 Free PMC article.
-
INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization.J Biol Chem. 2006 Sep 8;281(36):26754-67. doi: 10.1074/jbc.M604666200. Epub 2006 Jul 3. J Biol Chem. 2006. PMID: 16818491
-
Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction.J Biol Chem. 2013 Sep 13;288(37):26847-55. doi: 10.1074/jbc.M113.472415. Epub 2013 Aug 6. J Biol Chem. 2013. PMID: 23921379 Free PMC article.
-
Biophysics of actin filament severing by cofilin.FEBS Lett. 2013 Apr 17;587(8):1215-9. doi: 10.1016/j.febslet.2013.01.062. Epub 2013 Feb 5. FEBS Lett. 2013. PMID: 23395798 Free PMC article. Review.
-
Regulation of formin INF2 and its alteration in INF2-linked inherited disorders.Cell Mol Life Sci. 2024 Nov 25;81(1):463. doi: 10.1007/s00018-024-05499-3. Cell Mol Life Sci. 2024. PMID: 39586895 Free PMC article. Review.
Cited by
-
F-actin dismantling through a redox-driven synergy between Mical and cofilin.Nat Cell Biol. 2016 Aug;18(8):876-85. doi: 10.1038/ncb3390. Epub 2016 Jul 25. Nat Cell Biol. 2016. PMID: 27454820 Free PMC article.
-
3D electron tomographic and biochemical analysis of ER, Golgi and trans Golgi network membrane systems in stimulated Venus flytrap (Dionaea muscipula) glandular cells.J Biol Res (Thessalon). 2018 Aug 8;25:15. doi: 10.1186/s40709-018-0086-2. eCollection 2018 Dec. J Biol Res (Thessalon). 2018. PMID: 30116723 Free PMC article.
-
Mechanisms of formin-mediated actin assembly and dynamics.Biophys Rev. 2018 Dec;10(6):1553-1569. doi: 10.1007/s12551-018-0468-6. Epub 2018 Nov 3. Biophys Rev. 2018. PMID: 30392063 Free PMC article. Review.
-
Nanostructured self-assembly of inverted formin 2 (INF2) and F-actin-INF2 complexes revealed by atomic force microscopy.Langmuir. 2014 Jul 1;30(25):7533-9. doi: 10.1021/la501748x. Epub 2014 Jun 20. Langmuir. 2014. PMID: 24915113 Free PMC article.
-
FSGS-Causing INF2 Mutation Impairs Cleaved INF2 N-Fragment Functions in Podocytes.J Am Soc Nephrol. 2020 Feb;31(2):374-391. doi: 10.1681/ASN.2019050443. Epub 2020 Jan 10. J Am Soc Nephrol. 2020. PMID: 31924668 Free PMC article.
References
-
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

