Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting
- PMID: 24413018
- PMCID: PMC3989609
- DOI: 10.1002/embj.201386120
Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting
Abstract
For decades, neuroscientists have used enriched preparations of synaptic particles called synaptosomes to study synapse function. However, the interpretation of corresponding data is problematic as synaptosome preparations contain multiple types of synapses and non-synaptic neuronal and glial contaminants. We established a novel Fluorescence Activated Synaptosome Sorting (FASS) method that substantially improves conventional synaptosome enrichment protocols and enables high-resolution biochemical analyses of specific synapse subpopulations. Employing knock-in mice with fluorescent glutamatergic synapses, we show that FASS isolates intact ultrapure synaptosomes composed of a resealed presynaptic terminal and a postsynaptic density as assessed by light and electron microscopy. FASS synaptosomes contain bona fide glutamatergic synapse proteins but are almost devoid of other synapse types and extrasynaptic or glial contaminants. We identified 163 enriched proteins in FASS samples, of which FXYD6 and Tpd52 were validated as new synaptic proteins. FASS purification thus enables high-resolution biochemical analyses of specific synapse subpopulations in health and disease.
Figures



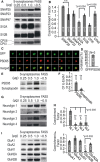

Following SDS-PAGE and tryptic digestion, proteins of both samples were analyzed by high-resolution tandem MS. The enrichment and depletion of proteins in sorted versus unsorted synaptosomes were determined by spectral counting using Scaffold. Of 1,075 quantified proteins, 163 were enriched by a factor of two or more, while 343 were depleted by a factor of two or more (listed in supplementary Tables S1–S3).
Fold change of protein spectral counts between the FASS-purified and S-synaptosomes for selected targets that were also analyzed by Western blotting (see Figs 4). Fold depletion is plotted as negative values and fold enrichment as positive values.
Spectral countings were compared with mRNA expression data of neurons, oligodendrocytes, and astrocytes (Cahoy et al, 2008). Relative contributions of genes with maximal expression in neurons (Nmax, NmaxA, NmaxO), astrocytes (Amax, AmaxN, AmaxO) and oligodendrocytes (Omax, OmaxN, OmaxA) are plotted as percentages of differentially expressed genes (see supplementary Fig S2 for detailed clusters). Among astrocyte and oligodendrocyte genes, percentages of genes classified to have relatively high expression in neurons are also plotted (e.g. AmaxN/(Amax+AmaxN+AmaxO); OmaxN/(Omax+OmaxN+OmaxA)).
Subcellular localizations of the 163 proteins enriched twofold or more in FASS samples.
Cellular functions of the 163 proteins enriched twofold or more in FASS samples.

References
-
- Abul-Husn NS, Devi LA. Neuroproteomics of the synapse and drug addiction. J Pharmacol Exp Ther. 2006;318:461–468. - PubMed
-
- Al-Hallaq RA, Yasuda RP, Wolfe BB. Enrichment of N-methyl-d-aspartate NR1 splice variants and synaptic proteins in rat postsynaptic densities. J Neurochem. 2001;77:110–119. - PubMed
-
- Bard L, Groc L. Glutamate receptor dynamics and protein interaction: lessons from the NMDA receptor. Mol Cell Neurosci. 2011;48:298–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

