CpG domains downstream of TSSs promote high levels of gene expression
- PMID: 24413563
- PMCID: PMC3973331
- DOI: 10.1093/nar/gkt1358
CpG domains downstream of TSSs promote high levels of gene expression
Abstract
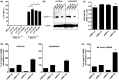
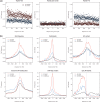
CpG dinucleotides are known to play a crucial role in regulatory domains, affecting gene expression in their natural context. Here, we demonstrate that intragenic CpG frequency and distribution impacts transgene and genomic gene expression levels in mammalian cells. As shown for the Macrophage Inflammatory Protein 1α, de novo RNA synthesis correlates with the number of CpG dinucleotides, whereas RNA splicing, stability, nuclear export and translation are not affected by the sequence modification. Differences in chromatin accessibility in vivo and altered nucleosome positioning in vitro suggest that increased CpG levels destabilize the chromatin structure. Moreover, enriched CpG levels correlate with increased RNA polymerase II elongation rates in vivo. Interestingly, elevated CpG levels particularly at the 5' end of the gene promote efficient transcription. We show that this is a genome-wide feature of highly expressed genes, by identifying a domain of ∼700 bp with high CpG content downstream of the transcription start site, correlating with high levels of transcription. We suggest that these 5' CpG domains are required to distort the chromatin structure and to increase gene activity.
Figures




References
-
- Welch M, Villalobos A, Gustafsson C, Minshull J. Designing genes for successful protein expression. Methods Enzymol. 2011;498:43–66. - PubMed
-
- Maertens B, Spriestersbach A, von Groll U, Roth U, Kubicek J, Gerrits M, Graf M, Liss M, Daubert D, Wagner R, et al. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci. Publ. Protein Soc. 2010;19:1312–1326. - PMC - PubMed
-
- Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

