Model organisms retain an "ecological memory" of complex ecologically relevant environmental variation
- PMID: 24413600
- PMCID: PMC3957629
- DOI: 10.1128/AEM.03280-13
Model organisms retain an "ecological memory" of complex ecologically relevant environmental variation
Abstract
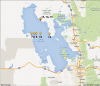
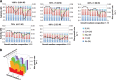
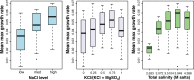
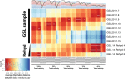
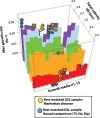
Although tractable model organisms are essential to characterize the molecular mechanisms of evolution and adaptation, the ecological relevance of their behavior is not always clear because certain traits are easily lost during long-term laboratory culturing. Here, we demonstrate that despite their long tenure in the laboratory, model organisms retain "ecological memory" of complex environmental changes. We have discovered that Halobacterium salinarum NRC-1, a halophilic archaeon that dominates microbial communities in a dynamically changing hypersaline environment, simultaneously optimizes fitness to total salinity, NaCl concentration, and the [K]/[Mg] ratio. Despite being maintained under controlled conditions over the last 50 years, peaks in the three-dimensional fitness landscape occur in salinity and ionic compositions that are not replicated in laboratory culturing but are routinely observed in the natural hypersaline environment of this organism. Intriguingly, adaptation to variations in ion composition was associated with differential regulation of anaerobic metabolism genes, suggesting an intertwined relationship between responses to oxygen and salinity. Our results suggest that the ecological memory of complex environmental variations is imprinted in the networks for coordinating multiple cellular processes. These coordination networks are also essential for dealing with changes in other physicochemically linked factors present during routine laboratory culturing and, hence, retained in model organisms.
Figures
References
-
- Oren A. 2001. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466:61–72. 10.1023/A:1014557116838 - DOI
-
- Tietge JE, Hockett JR, Evans JM. 1997. Major ion toxicity of six produced waters to three freshwater species: application of ion toxicity models and TIE procedures. Environ. Toxicol. Chem. 16:2002–2008. 10.1002/etc.5620161004 - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

