Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas
- PMID: 24413734
- PMCID: PMC3963408
- DOI: 10.1038/ng.2873
Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas
Abstract
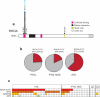
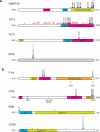
Peripheral T cell lymphomas (PTCLs) are a heterogeneous and poorly understood group of non-Hodgkin lymphomas. Here we combined whole-exome sequencing of 12 tumor-normal DNA pairs, RNA sequencing analysis and targeted deep sequencing to identify new genetic alterations in PTCL transformation. These analyses identified highly recurrent epigenetic factor mutations in TET2, DNMT3A and IDH2 as well as a new highly prevalent RHOA mutation encoding a p.Gly17Val alteration present in 22 of 35 (67%) angioimmunoblastic T cell lymphoma (AITL) samples and in 8 of 44 (18%) PTCL, not otherwise specified (PTCL-NOS) samples. Mechanistically, the RHOA Gly17Val protein interferes with RHOA signaling in biochemical and cellular assays, an effect potentially mediated by the sequestration of activated guanine-exchange factor (GEF) proteins. In addition, we describe new and recurrent, albeit less frequent, genetic defects including mutations in FYN, ATM, B2M and CD58 implicating SRC signaling, impaired DNA damage response and escape from immune surveillance mechanisms in the pathogenesis of PTCL.
Figures


References
-
- Armitage JO. The aggressive peripheral T-cell lymphomas: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2012;87:511–9. - PubMed
-
- Rudiger T, et al. Peripheral T-cell lymphoma (excluding anaplastic large-cell lymphoma): results from the Non-Hodgkin's Lymphoma Classification Project. Ann Oncol. 2002;13:140–9. - PubMed
-
- Schiller MR. Coupling receptor tyrosine kinases to Rho GTPases--GEFs what's the link. Cell Signal. 2006;18:1834–43. - PubMed
-
- Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–38. - PubMed
-
- Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

