RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia
- PMID: 24413735
- PMCID: PMC3960636
- DOI: 10.1038/ng.2874
RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia
Abstract
The ETV6-RUNX1 fusion gene, found in 25% of childhood acute lymphoblastic leukemia (ALL) cases, is acquired in utero but requires additional somatic mutations for overt leukemia. We used exome and low-coverage whole-genome sequencing to characterize secondary events associated with leukemic transformation. RAG-mediated deletions emerge as the dominant mutational process, characterized by recombination signal sequence motifs near breakpoints, incorporation of non-templated sequence at junctions, ∼30-fold enrichment at promoters and enhancers of genes actively transcribed in B cell development and an unexpectedly high ratio of recurrent to non-recurrent structural variants. Single-cell tracking shows that this mechanism is active throughout leukemic evolution, with evidence of localized clustering and reiterated deletions. Integration of data on point mutations and rearrangements identifies ATF7IP and MGA as two new tumor-suppressor genes in ALL. Thus, a remarkably parsimonious mutational process transforms ETV6-RUNX1-positive lymphoblasts, targeting the promoters, enhancers and first exons of genes that normally regulate B cell differentiation.
Figures



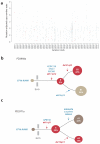
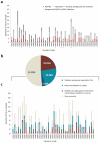
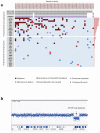

Comment in
-
A RAG driver on the road to pediatric ALL.Nat Genet. 2014 Feb;46(2):96-8. doi: 10.1038/ng.2879. Nat Genet. 2014. PMID: 24473322
References
-
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nature reviews. Cancer. 2003;3:639–649. - PubMed
-
- Bateman CM, et al. Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood. 2010;115:3553–3558. - PubMed
-
- Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 101880/WT_/Wellcome Trust/United Kingdom
- WT100183MA/WT_/Wellcome Trust/United Kingdom
- 088340/WT_/Wellcome Trust/United Kingdom
- R01 CA172558/CA/NCI NIH HHS/United States
- R01 CA169458/CA/NCI NIH HHS/United States
- 077012/05/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA139032/CA/NCI NIH HHS/United States
- 18131/CRUK_/Cancer Research UK/United Kingdom
- R01 CA137060/CA/NCI NIH HHS/United States
- R01 CA157644/CA/NCI NIH HHS/United States
- WT088340MA/WT_/Wellcome Trust/United Kingdom
- 093867/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

