Active DNA demethylation is required for complete imprint erasure in primordial germ cells
- PMID: 24413819
- PMCID: PMC3888974
- DOI: 10.1038/srep03658
Active DNA demethylation is required for complete imprint erasure in primordial germ cells
Abstract
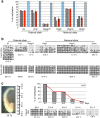
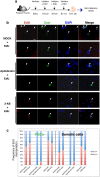
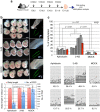
In mammalian primordial germ cells (PGCs), DNA demethylation is indispensible for parental imprint erasure, which is a reprogramming process essential for normal developmental potential. Thus, it is important to elucidate how DNA demethylation occurs in each imprinted region in PGCs and to determine which DNA demethylation pathway, passive or active, essentially contributes to the erasure of the imprint. Here, we report that active DNA demethylation via a putative Poly(ADP-ribose) polymerase (PARP) pathway is involved in H19-DMR imprint erasure in PGCs, as shown by an in vivo small molecule inhibitor assay. To the best of our knowledge, this is the first direct demonstration of a DNA replication-independent active DNA demethylation pathway in the erasure process of genomic imprinting in PGCs in vivo. The data also suggest that active DNA demethylation plays a significant role in the complete erasure of paternal imprinting in the female germ line.
Figures
Similar articles
-
Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):9545-50. doi: 10.1073/pnas.1610259113. Epub 2016 Aug 2. Proc Natl Acad Sci U S A. 2016. PMID: 27486249 Free PMC article.
-
Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles.PLoS One. 2012;7(10):e46927. doi: 10.1371/journal.pone.0046927. Epub 2012 Oct 5. PLoS One. 2012. PMID: 23071665 Free PMC article.
-
Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine.Science. 2013 Jan 25;339(6118):448-52. doi: 10.1126/science.1229277. Epub 2012 Dec 6. Science. 2013. PMID: 23223451 Free PMC article.
-
Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells.Curr Opin Cell Biol. 2013 Jun;25(3):281-8. doi: 10.1016/j.ceb.2013.02.013. Epub 2013 Mar 17. Curr Opin Cell Biol. 2013. PMID: 23510682 Review.
-
Genomic imprinting is a parental effect established in mammalian germ cells.Curr Top Dev Biol. 2013;102:35-59. doi: 10.1016/B978-0-12-416024-8.00002-7. Curr Top Dev Biol. 2013. PMID: 23287029 Review.
Cited by
-
Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):9545-50. doi: 10.1073/pnas.1610259113. Epub 2016 Aug 2. Proc Natl Acad Sci U S A. 2016. PMID: 27486249 Free PMC article.
-
Mammalian-specific genomic functions: Newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals.Proc Jpn Acad Ser B Phys Biol Sci. 2015;91(10):511-38. doi: 10.2183/pjab.91.511. Proc Jpn Acad Ser B Phys Biol Sci. 2015. PMID: 26666304 Free PMC article. Review.
-
Mechanisms underlying low mutation rates in mammalian oocytes and preimplantation embryos.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf760. doi: 10.1093/nar/gkaf760. Nucleic Acids Res. 2025. PMID: 40795959 Free PMC article.
-
Targeted DNA methylation in pericentromeres with genome editing-based artificial DNA methyltransferase.PLoS One. 2017 May 18;12(5):e0177764. doi: 10.1371/journal.pone.0177764. eCollection 2017. PLoS One. 2017. PMID: 28542388 Free PMC article.
-
Are there specific readers of oxidized 5-methylcytosine bases?Bioessays. 2016 Oct;38(10):1038-47. doi: 10.1002/bies.201600126. Epub 2016 Aug 2. Bioessays. 2016. PMID: 27480808 Free PMC article. Review.
References
-
- Surani M. A., Barton S. C. & Noris M. L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550 (1984). - PubMed
-
- McGrath J. & Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 (1984). - PubMed
-
- Cattanach B. M. & Kirk M. Differential activity of maternally and paternally derived chromosome regions in mouse. Nature 315, 496–498 (1985). - PubMed
-
- Reik W., Dean W. & Walter J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

