EWS-FLI-1 regulates the neuronal repressor gene REST, which controls Ewing sarcoma growth and vascular morphology
- PMID: 24415532
- PMCID: PMC3969734
- DOI: 10.1002/cncr.28555
EWS-FLI-1 regulates the neuronal repressor gene REST, which controls Ewing sarcoma growth and vascular morphology
Abstract
Background: RE1-silencing transcription factor (REST), a neuronal repressor gene, regulates neuronal stem cell differentiation. Ewing sarcoma may originate from neural crest cells. In the current study, the authors investigated whether REST plays a role in the growth of this tumor.
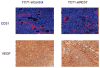
Methods: REST expression was determined by Western blot analysis and reverse transcription-polymerase chain reaction in 3 human Ewing sarcoma cell lines and 7 patient tumor samples. The role of REST in tumor growth and tumor vascular morphology was determined using a Ewing sarcoma xenograft model. Immunofluorescence staining, Hypoxyprobe, and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assays were performed to investigate the impact of REST on pericyte marker expression, hypoxia, and apoptosis in vivo.
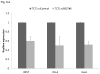
Results: High levels of REST were expressed in all 3 human Ewing sarcoma cell lines and in 6 of the 7 patient tumor samples. Overexpression of EWS-FLI-1 in human mesenchymal stem cells and human neural progenitor cells was found to increase REST expression. Inhibition of EWS-FLI-1 using small interfering RNA decreased REST expression in human Ewing sarcoma cells. Inhibition of REST did not affect EWS-FLI-1, but significantly suppressed tumor growth in vivo, reduced the tumor vessel pericyte markers α- smooth muscle actin (SMA) and desmin, increased hypoxia and apoptosis in tumor tissues, and decreased the expression of delta-like ligand 4 (DLL4) and Hes1.
Conclusions: Inhibition of REST suppressed tumor growth, inhibited pericyte marker expression, and increased tumor hypoxia and apoptosis. Because tumor vessel function has been linked to tumor growth and metastases, REST may be a new therapeutic target in patients with Ewing sarcoma.
Keywords: EWS-FLI-1; Ewing sarcoma; RE1-silencing transcription factor (REST); tumor growth; tumor vasculature.
© 2013 American Cancer Society.
Conflict of interest statement
Conflict of interest disclosures: All authors declare no financial disclosure.
Figures





















References
-
- Riggi N, Stamenkovic I. The Biology of Ewing sarcoma. Cancer Lett. 2007;254:1–10. - PubMed
-
- Barker LM, Pendergrass TW, Sanders JE, Hawkins DS. Survival after recurrence of Ewing's sarcoma family of tumors. J Clin Oncol. 2005;23:4354–4362. - PubMed
-
- Miser JS, Krailo MD, Tarbell NJ, et al. Treatment of metastatic Ewing's sarcoma or primitive neuroectodermal tumor of bone: evaluation of combination ifosfamide and etoposide--a Children's Cancer Group and Pediatric Oncology Group study. J Clin Oncol. 2004;22:2873–2876. - PubMed
-
- Rorie CJ, Thomas VD, Chen P, Pierce HH, O'Bryan JP, Weissman BE. The Ews/Fli-1 fusion gene switches the differentiation program of neuroblastomas to Ewing sarcoma/peripheral primitive neuroectodermal tumors. Cancer Res. 2004;64:1266–1277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

