Characterization and molecular profiling of PSEN1 familial Alzheimer's disease iPSC-derived neural progenitors
- PMID: 24416243
- PMCID: PMC3885572
- DOI: 10.1371/journal.pone.0084547
Characterization and molecular profiling of PSEN1 familial Alzheimer's disease iPSC-derived neural progenitors
Abstract
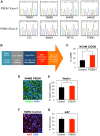
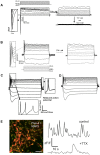
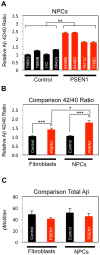
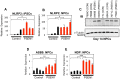
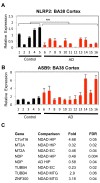
Presenilin 1 (PSEN1) encodes the catalytic subunit of γ-secretase, and PSEN1 mutations are the most common cause of early onset familial Alzheimer's disease (FAD). In order to elucidate pathways downstream of PSEN1, we characterized neural progenitor cells (NPCs) derived from FAD mutant PSEN1 subjects. Thus, we generated induced pluripotent stem cells (iPSCs) from affected and unaffected individuals from two families carrying PSEN1 mutations. PSEN1 mutant fibroblasts, and NPCs produced greater ratios of Aβ42 to Aβ40 relative to their control counterparts, with the elevated ratio even more apparent in PSEN1 NPCs than in fibroblasts. Molecular profiling identified 14 genes differentially-regulated in PSEN1 NPCs relative to control NPCs. Five of these targets showed differential expression in late onset AD/Intermediate AD pathology brains. Therefore, in our PSEN1 iPSC model, we have reconstituted an essential feature in the molecular pathogenesis of FAD, increased generation of Aβ42/40, and have characterized novel expression changes.
Conflict of interest statement
Figures

References
-
- Quintero-Monzon O, Martin MM, Fernandez MA, Cappello CA, Krzysiak AJ, et al. (2011) Dissociation between the processivity and total activity of γ-secretase: implications for the mechanism of Alzheimer's disease-causing presenilin mutations. Biochemistry 50: 9023–9035 10.1021/bi2007146 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K12 GM068524/GM/NIGMS NIH HHS/United States
- P01AG07232/AG/NIA NIH HHS/United States
- R01 MH091844/MH/NIMH NIH HHS/United States
- R01 NS049442/NS/NINDS NIH HHS/United States
- R01AG037212/AG/NIA NIH HHS/United States
- R01MH091844/MH/NIMH NIH HHS/United States
- P50AG08702/AG/NIA NIH HHS/United States
- R21AG042965/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- P01 AG007232/AG/NIA NIH HHS/United States
- RF1 AG042965/AG/NIA NIH HHS/United States
- 1U01AG046170-01/AG/NIA NIH HHS/United States
- NS049442/NS/NINDS NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- R01 AG037212/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

