The SARS coronavirus nucleocapsid protein--forms and functions
- PMID: 24418573
- PMCID: PMC7113676
- DOI: 10.1016/j.antiviral.2013.12.009
The SARS coronavirus nucleocapsid protein--forms and functions
Abstract
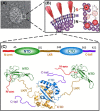
The nucleocapsid phosphoprotein of the severe acute respiratory syndrome coronavirus (SARS-CoV N protein) packages the viral genome into a helical ribonucleocapsid (RNP) and plays a fundamental role during viral self-assembly. It is a protein with multifarious activities. In this article we will review our current understanding of the N protein structure and its interaction with nucleic acid. Highlights of the progresses include uncovering the modular organization, determining the structures of the structural domains, realizing the roles of protein disorder in protein-protein and protein-nucleic acid interactions, and visualizing the ribonucleoprotein (RNP) structure inside the virions. It was also demonstrated that N-protein binds to nucleic acid at multiple sites with a coupled-allostery manner. We propose a SARS-CoV RNP model that conforms to existing data and bears resemblance to the existing RNP structures of RNA viruses. The model highlights the critical role of modular organization and intrinsic disorder of the N protein in the formation and functions of the dynamic RNP capsid in RNA viruses. This paper forms part of a symposium in Antiviral Research on "From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses."
Keywords: Capsid packaging; Coronavirus; Intrinsic disorder; Nucleocapsid protein; RNP; SARS.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures



Comment in
-
The Nucleocapsid Protein of SARS-CoV-2: a Target for Vaccine Development.J Virol. 2020 Jun 16;94(13):e00647-20. doi: 10.1128/JVI.00647-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32546606 Free PMC article. No abstract available.
References
-
- Ababou A., Ladbury J.E. Survey of the year 2005: literature on applications of isothermal titration calorimetry. J. Mol. Recognit. 2007;20:4–14. - PubMed
-
- Albertini A.A.V., Wernimont A.K., Muziol T., Ravelli R.B.G., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W.H. Crystal structure of the rabies virus nucleoprotein–RNA complex. Science. 2006;313:360–363. - PubMed
-
- Ariza A., Tanner S.J., Walter C.T., Dent K.C., Shepherd D.A., Wu W.N., Matthews S.V., Hiscox J.A., Green T.J., Luo M., Elliott R.M., Fooks A.R., Ashcroft A.E., Stonehouse N.J., Ranson N.A., Barr J.N., Edwards T.A. Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization. Nucleic Acids Res.2013;41:5912–5926. - PMC - PubMed
-
- Caul E.O., Egglestone S.I. Coronavirus-like particles present in simian faeces. Vet. Rec. 1979;104:168–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

