Reconstructing complex regions of genomes using long-read sequencing technology
- PMID: 24418700
- PMCID: PMC3975067
- DOI: 10.1101/gr.168450.113
Reconstructing complex regions of genomes using long-read sequencing technology
Abstract
Obtaining high-quality sequence continuity of complex regions of recent segmental duplication remains one of the major challenges of finishing genome assemblies. In the human and mouse genomes, this was achieved by targeting large-insert clones using costly and laborious capillary-based sequencing approaches. Sanger shotgun sequencing of clone inserts, however, has now been largely abandoned, leaving most of these regions unresolved in newer genome assemblies generated primarily by next-generation sequencing hybrid approaches. Here we show that it is possible to resolve regions that are complex in a genome-wide context but simple in isolation for a fraction of the time and cost of traditional methods using long-read single molecule, real-time (SMRT) sequencing and assembly technology from Pacific Biosciences (PacBio). We sequenced and assembled BAC clones corresponding to a 1.3-Mbp complex region of chromosome 17q21.31, demonstrating 99.994% identity to Sanger assemblies of the same clones. We targeted 44 differences using Illumina sequencing and find that PacBio and Sanger assemblies share a comparable number of validated variants, albeit with different sequence context biases. Finally, we targeted a poorly assembled 766-kbp duplicated region of the chimpanzee genome and resolved the structure and organization for a fraction of the cost and time of traditional finishing approaches. Our data suggest a straightforward path for upgrading genomes to a higher quality finished state.
Figures
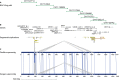
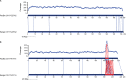


Comment in
-
Technology: SMRT move?Nat Rev Genet. 2014 Mar;15(3):146. doi: 10.1038/nrg3678. Epub 2014 Feb 4. Nat Rev Genet. 2014. PMID: 24492234 No abstract available.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- SRA/SRP035602
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
