The structure of endothiapepsin complexed with a Phe-Tyr reduced-bond inhibitor at 1.35 Å resolution
- PMID: 24419612
- PMCID: PMC3943106
- DOI: 10.1107/S2053230X13032974
The structure of endothiapepsin complexed with a Phe-Tyr reduced-bond inhibitor at 1.35 Å resolution
Abstract
Endothiapepsin is a typical member of the aspartic proteinase family. The catalytic mechanism of this family is attributed to two conserved catalytic aspartate residues, which coordinate the hydrolysis of a peptide bond. An oligopeptide inhibitor (IC50 = 0.62 µM) based on a reduced-bond transition-state inhibitor of mucorpepsin was co-crystallized with endothiapepsin and the crystal structure of the enzyme-inhibitor complex was determined at 1.35 Å resolution. A total of 12 hydrogen bonds between the inhibitor and the active-site residues were identified. The resulting structure demonstrates a number of novel subsite interactions in the active-site cleft.
Keywords: aspartic proteases; endothiapepsin; inhibitor; transition state.
Figures
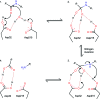

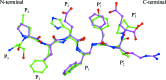
 Tyr-Thr-Pro. There is a reduced peptide bond (indicated by an asterisk) between Phe and Tyr, i.e. the carbonyl group is replaced by a methylene –CH2– moiety.
Tyr-Thr-Pro. There is a reduced peptide bond (indicated by an asterisk) between Phe and Tyr, i.e. the carbonyl group is replaced by a methylene –CH2– moiety.


Similar articles
-
X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.J Mol Biol. 1990 Dec 20;216(4):1017-29. doi: 10.1016/S0022-2836(99)80017-5. J Mol Biol. 1990. PMID: 2266553
-
X-ray, neutron and NMR studies of the catalytic mechanism of aspartic proteinases.Eur Biophys J. 2006 Sep;35(7):559-66. doi: 10.1007/s00249-006-0065-7. Epub 2006 May 4. Eur Biophys J. 2006. PMID: 16673078
-
Atomic resolution analysis of the catalytic site of an aspartic proteinase and an unexpected mode of binding by short peptides.Protein Sci. 2003 Aug;12(8):1741-9. doi: 10.1110/ps.0305203. Protein Sci. 2003. PMID: 12876323 Free PMC article.
-
Aspartic proteases in drug discovery.Curr Pharm Des. 2007;13(3):271-85. doi: 10.2174/138161207779313560. Curr Pharm Des. 2007. PMID: 17313361 Review.
-
On the active site protonation state in aspartic proteases: implications for drug design.Curr Pharm Des. 2013;19(23):4257-75. doi: 10.2174/1381612811319230009. Curr Pharm Des. 2013. PMID: 23170891 Review.
References
-
- Bailey, D. et al. (2012). Acta Cryst. D68, 541–552. - PubMed
-
- Blundell, T. L., Jenkins, J. A., Sewell, B. T., Pearl, L. H., Cooper, J. B., Tickle, I. J., Veerapandian, B. & Wood, S. P. (1990). J. Mol. Biol. 211, 919–941. - PubMed
-
- Coates, L., Erskine, P. T., Crump, M. P., Wood, S. P. & Cooper, J. B. (2002). J. Mol. Biol. 318, 1405–1415. - PubMed
-
- Dhanaraj, V. et al. (1992). Nature (London), 357, 466–472. - PubMed
-
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

