Role of the C-terminal region of vervet monkey polyomavirus 1 VP1 in virion formation
- PMID: 24419975
- PMCID: PMC4073331
- DOI: 10.1292/jvms.13-0568
Role of the C-terminal region of vervet monkey polyomavirus 1 VP1 in virion formation
Abstract
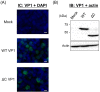
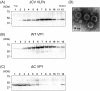
Recently, we detected novel vervet monkey polyomavirus 1 (VmPyV) in a vervet monkey. Among amino acid sequences of major capsid protein VP1s of other polyomaviruses, VmPyV VP1 is the longest with additional amino acid residues in the C-terminal region. To examine the role of VmPyV VP1 in virion formation, we generated virus-like particles (VLPs) of VmPyV VP1, because VLP is a useful tool for the investigation of the morphological characters of polyomavirus virions. After the full-length VmPyV VP1 was subcloned into a mammalian expression plasmid, the plasmid was transfected into human embryonic kidney 293T (HEK293T) cells. Thereafter, VmPyV VLPs were purified from the cell lysates of the transfected cells via sucrose gradient sedimentation. Electron microscopic analyses revealed that VmPyV VP1 forms VLPs with a diameter of approximately 50 nm that are exclusively localized in cell nuclei. Furthermore, we generated VLPs consisting of the deletion mutant VmPyV VP1 (ΔC VP1) lacking the C-terminal 116 amino acid residues and compared its VLP formation efficiency and morphology to those of VLPs from wild-type VmPyV VP1 (WT VP1). WT and ΔC VP1 VLPs were similar in size, but the number of ΔC VP1 VLPs was much lower than that of WT VP1 VLPs in VP1-expressing HEK293T cells. These results suggest that the length of VP1 is unrelated to virion morphology; however, the C-terminal region of VmPyV VP1 affects the efficiency of its VLP formation.
Figures


Similar articles
-
Hamster polyomavirus-derived virus-like particles are able to transfer in vitro encapsidated plasmid DNA to mammalian cells.Virus Genes. 2007 Jun;34(3):303-14. doi: 10.1007/s11262-006-0028-1. Epub 2006 Aug 22. Virus Genes. 2007. PMID: 16927120
-
Production of recombinant VP1-derived virus-like particles from novel human polyomaviruses in yeast.BMC Biotechnol. 2015 Aug 4;15:68. doi: 10.1186/s12896-015-0187-z. BMC Biotechnol. 2015. PMID: 26239840 Free PMC article.
-
Purification of recombinant trichodysplasia spinulosa-associated polyomavirus VP1-derived virus-like particles using chromatographic techniques.J Chromatogr B Analyt Technol Biomed Life Sci. 2018 Jul 15;1090:7-13. doi: 10.1016/j.jchromb.2018.05.007. Epub 2018 May 9. J Chromatogr B Analyt Technol Biomed Life Sci. 2018. PMID: 29778875
-
[Epidemiological and basic research activity targeting polyomaviruses].Uirusu. 2014;64(1):25-34. doi: 10.2222/jsv.64.25. Uirusu. 2014. PMID: 25765977 Review. Japanese.
-
Immunotherapeutic polyoma and human papilloma virus-like particles.Immunotherapy. 2009 Mar;1(2):303-12. doi: 10.2217/1750743X.1.2.303. Immunotherapy. 2009. PMID: 20635947 Review.
References
-
- Anthony S. J., St Leger J. A., Navarrete-Macias I., Nilson E., Sanchez-Leon M., Liang E., Seimon T., Jain K., Karesh W., Daszak P., Briese T., Lipkin W. I.2013. Identification of a novel cetacean polyomavirus from a common dolphin (Delphinus delphis) with Tracheobronchitis. PLoS ONE 8: e68239. doi: 10.1371/journal.pone.0068239 - DOI - PMC - PubMed
-
- Chang D., Fung C. Y., Ou W. C., Chao P. C., Li S. Y., Wang M., Huang Y. L., Tzeng T. Y., Tsai R. T.1997. Self-assembly of the JC virus major capsid protein, VP1, expressed in insect cells. J. Gen. Virol. 78: 1435–1439 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

