A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface
- PMID: 24421336
- PMCID: PMC3958310
- DOI: 10.1002/emmm.201303404
A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface
Abstract
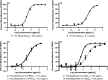
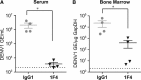
Dengue virus (DENV), which consists of four serotypes (DENV1-4), infects over 400 million people annually. Previous studies have indicated most human monoclonal antibodies (HMAbs) from dengue patients are cross-reactive and poorly neutralizing. Rare neutralizing HMAbs are usually serotype-specific and bind to quaternary structure-dependent epitopes. We determined the structure of DENV1 complexed with Fab fragments of a highly potent HMAb 1F4 to 6 Å resolution by cryo-EM. Although HMAb 1F4 appeared to bind to virus and not E proteins in ELISAs in the previous study, our structure showed that the epitope is located within an envelope (E) protein monomer, and not across neighboring E proteins. The Fab molecules bind to domain I (DI), and DI-DII hinge of the E protein. We also showed that HMAb 1F4 can neutralize DENV at different stages of viral entry in a cell type and receptor dependent manner. The structure reveals the mechanism by which this potent and specific antibody blocks viral infection.
Figures

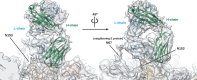
Cryo-EM map of Fab 1F4 complexed with DENV1 showed 120 copies of Fab (blue) bound to the virus surface (cyan). White triangle indicates an icosahedral asymmetric unit and the numbers represents the vertices.
Cross-section of a quarter of a cryo-EM map.
The resolution of the cryoEM map is 6 Å. Regions of the density map corresponding to trans-membrane α-helices (left) and β-strands (right).
The density map corresponding to Fab 1F4. The density of the constant region (indicated by arrow) is much poorer than the variable region indicating the constant region is flexible.

Densities of Fab 1F4 on E proteins in two icosahedral asymmetric units. Fab 1F4 molecules bind to molecules A and B in each asymmetric unit.
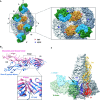
Open-book representation of Fab 1F4 binding to E protein. The E protein DI, DII and DIII are colored in red, yellow and blue (top), respectively, whereas the Fab 1F4 heavy and light chains are colored in green and cyan (bottom), respectively. The epitope bound by Fab 1F4 is located on the DI and DI-II hinge regions (top). The epitope on E protein bound by the heavy and light chain are colored in green and cyan, respectively (top). Also the epitope on DI that interacted with both heavy and light chains are colored in purple (top). The paratope on heavy and light chain (bottom) are colored in its corresponding interacting residues on the DI (red) and DII (yellow) E protein, paratope binding to both DI and II on the E protein is colored in pink.
Ribbon representation of the HMAb 1F4 epitope on E protein (top) and also sequence alignment of the epitope region between all dengue virus serotypes (bottom). The β-strand and loop of the epitope are colored in orange and black, respectively (top). In the bottom panel, the secondary structures are shown above the amino acid sequence. The amino acid residues that interact with Fab 1F4 are highlighted in the same coloring scheme as in (B). The amino acid sequences are derived from DENV1 strain PVP159, DENV2 strain S16803, DENV3 strain Thailand 1995 and DENV4 strain Dominica 1981.

Open-book representation showing the electrostatic potential of the interaction interface on the E protein (left) and the Fab 1F4 (right). The blue and red colors indicate positive and negative charges, respectively, whereas the white color shows neutral charge. The dotted lines indicate the border of the footprint between heavy and light chains on the E protein epitope (left) and also the corresponding border on the antibody paratope (right). Residues on E protein identified in the neutralization escape mutants (K47 and G274) are indicated, together with other residues that have opposite charges in other serotypes. The corresponding interacting residues on the Fab 1F4 light and heavy chains are indicated with cyan and green colored fonts, respectively.
Table of the list of putative interactions between DENV1 and Fab 1F4.



Superposition of the crystal structures of rE and the subnanometer resolution cryo-EM structures of virus E proteins showed that the hinge angles of rE proteins are variable whereas those on the virus surface are largely conserved. DI of these structures was superimposed using ‘LSQ superpose’ in COOT (Emsley et al, 2010).
Zoom-in view of the part of the HMAb 1F4 epitope that consists of the kl loop (represented as sticks) in the DI-II hinge region. The kl loops from the viruses (left) showed conserved conformation whereas those from the rE proteins (right) showed varying conformations. Residue 274, which is critical for HMAb 1F4 binding (De Alwis et al, 2012), is shown as a sphere.
HMAb 1F4 may block DC-SIGN from binding to the glycosylation site on N67. One DC-SIGN molecule binds across two N67 glycans on the virus surface. When Fab 1F4 is allowed to bind to the virus surface, it engages the N67 glycan on molecule B. This would cause steric hindrance thereby preventing the virus from binding to DC-SIGN on the surface of dendritic cells (Pokidysheva et al, 2006). Superposition of the cryo-EM complex structures of DENV1-Fab 1F4 and the DENV2-carbohydrate recognition domain (CRD) of DC-SIGN (PDB code 2B6B) showed that the Fab and the CRD molecules clashed. This indicates that simultaneous binding of these molecules on the virus surface is not possible. E protein molecules A, B and C are colored in light gray and molecules A’, B’ and C’ in gray. DC-SIGN is shown as a transparent orange surface. The glycosylation sites on N67 and N153 are marked as red stars and blue squares, respectively. The clashes between the Fab and DC-SIGN molecules are indicated by arrows.
Fab 1F4 may interfere with the E protein dimer to trimer structural changes during fusion of the virus to the endosomal membrane. (i) The DI-DII hinge of the whole virus E protein has a different angle compared to the crystal structure of the trimeric post- fusion rE protein. This suggests that the binding of HMAb 1F4 to virus may lock the E protein hinge thus preventing the kl loop from undergoing structural changes to the post-fusion structure. The E protein on the virus and the post-fusion trimeric state are colored in blue and pink, respectively. (ii) Superposition of the cryo-EM Fab 1F4-DENV1 E protein structure onto the post-fusion trimeric E protein crystal structure. The Fab 1F4 molecule clashed with DIII of a neighboring E protein in the post-fusion trimeric structure (indicated by arrow). Two molecules of E protein are shown as surfaces (colored in light gray and dark gray) and one molecule is shown as ribbons. Fab 1F4 is drawn as a transparent surface.
References
-
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
-
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

