Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation
- PMID: 24421395
- PMCID: PMC3958034
- DOI: 10.1128/MCB.01372-13
Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation
Abstract
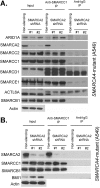
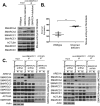
Collectively, genes encoding subunits of the SWI/SNF (BAF) chromatin remodeling complex are mutated in 20% of all human cancers, with the SMARCA4 (BRG1) subunit being one of the most frequently mutated. The SWI/SNF complex modulates chromatin remodeling through the activity of two mutually exclusive catalytic subunits, SMARCA4 and SMARCA2 (BRM). Here, we show that a SMARCA2-containing residual SWI/SNF complex underlies the oncogenic activity of SMARCA4 mutant cancers. We demonstrate that a residual SWI/SNF complex exists in SMARCA4 mutant cell lines and plays essential roles in cellular proliferation. Further, using data from loss-of-function screening of 165 cancer cell lines, we identify SMARCA2 as an essential gene in SMARCA4 mutant cancer cell lines. Mechanistically, we reveal that Smarca4 inactivation leads to greater incorporation of the nonessential SMARCA2 subunit into the SWI/SNF complex. Collectively, these results reveal a role for SMARCA2 in oncogenesis caused by SMARCA4 loss and identify the ATPase and bromodomain-containing SMARCA2 as a potential therapeutic target in these cancers.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
