The skin microbiome in healthy and allergic dogs
- PMID: 24421875
- PMCID: PMC3885435
- DOI: 10.1371/journal.pone.0083197
The skin microbiome in healthy and allergic dogs
Abstract
Background: Changes in the microbial populations on the skin of animals have traditionally been evaluated using conventional microbiology techniques. The sequencing of bacterial 16S rRNA genes has revealed that the human skin is inhabited by a highly diverse and variable microbiome that had previously not been demonstrated by culture-based methods. The goals of this study were to describe the microbiome inhabiting different areas of the canine skin, and to compare the skin microbiome of healthy and allergic dogs.
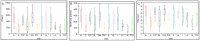
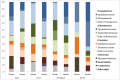
Methodology/principal findings: DNA extracted from superficial skin swabs from healthy (n = 12) and allergic dogs (n = 6) from different regions of haired skin and mucosal surfaces were used for 454-pyrosequencing of the 16S rRNA gene. Principal coordinates analysis revealed clustering for the different skin sites across all dogs, with some mucosal sites and the perianal regions clustering separately from the haired skin sites. The rarefaction analysis revealed high individual variability between samples collected from healthy dogs and between the different skin sites. Higher species richness and microbial diversity were observed in the samples from haired skin when compared to mucosal surfaces or mucocutaneous junctions. In all examined regions, the most abundant phylum and family identified in the different regions of skin and mucosal surfaces were Proteobacteria and Oxalobacteriaceae. The skin of allergic dogs had lower species richness when compared to the healthy dogs. The allergic dogs had lower proportions of the Betaproteobacteria Ralstonia spp. when compared to the healthy dogs.
Conclusions/significance: The study demonstrates that the skin of dogs is inhabited by much more rich and diverse microbial communities than previously thought using culture-based methods. Our sequence data reveal high individual variability between samples collected from different patients. Differences in species richness was also seen between healthy and allergic dogs, with allergic dogs having lower species richness when compared to healthy dogs.
Conflict of interest statement
Figures





Similar articles
-
Description and comparison of the skin and ear canal microbiota of non-allergic and allergic German shepherd dogs using next generation sequencing.PLoS One. 2021 May 3;16(5):e0250695. doi: 10.1371/journal.pone.0250695. eCollection 2021. PLoS One. 2021. PMID: 33939741 Free PMC article.
-
Bacterial microbiome in the nose of healthy cats and in cats with nasal disease.PLoS One. 2017 Jun 29;12(6):e0180299. doi: 10.1371/journal.pone.0180299. eCollection 2017. PLoS One. 2017. PMID: 28662139 Free PMC article.
-
The feline skin microbiota: The bacteria inhabiting the skin of healthy and allergic cats.PLoS One. 2017 Jun 2;12(6):e0178555. doi: 10.1371/journal.pone.0178555. eCollection 2017. PLoS One. 2017. PMID: 28575016 Free PMC article.
-
Current state of knowledge: the canine gastrointestinal microbiome.Anim Health Res Rev. 2012 Jun;13(1):78-88. doi: 10.1017/S1466252312000059. Epub 2012 May 30. Anim Health Res Rev. 2012. PMID: 22647637 Review.
-
The Microbiota Regulates Immunity and Immunologic Diseases in Dogs and Cats.Vet Clin North Am Small Anim Pract. 2018 Mar;48(2):307-322. doi: 10.1016/j.cvsm.2017.10.008. Epub 2017 Nov 29. Vet Clin North Am Small Anim Pract. 2018. PMID: 29198905 Review.
Cited by
-
Characterization of skin surface and dermal microbiota in dogs with mast cell tumor.Sci Rep. 2020 Jul 28;10(1):12634. doi: 10.1038/s41598-020-69572-0. Sci Rep. 2020. PMID: 32724217 Free PMC article.
-
Unenriched skin microbiota transplantation for cats: new road story for treating feline atopic skin syndrome.Braz J Vet Med. 2023 Dec 2;45:e002823. doi: 10.29374/2527-2179.bjvm002823. eCollection 2023. Braz J Vet Med. 2023. PMID: 38093985 Free PMC article.
-
Comparison of hybrid learning and remote education in the implementation of the "Adopt a Microorganism" methodology.PLoS One. 2021 Nov 24;16(11):e0248906. doi: 10.1371/journal.pone.0248906. eCollection 2021. PLoS One. 2021. PMID: 34818328 Free PMC article.
-
Quantification of Malassezia pachydermatis by real-time PCR in swabs from the external ear canal of dogs.J Vet Diagn Invest. 2019 May;31(3):440-447. doi: 10.1177/1040638719840686. Epub 2019 Apr 4. J Vet Diagn Invest. 2019. PMID: 30943876 Free PMC article.
-
Influence of clipping on bacterial contamination of canine arthrocentesis sites before and after skin preparation.Vet Surg. 2020 Oct;49(7):1307-1314. doi: 10.1111/vsu.13468. Epub 2020 Jun 9. Vet Surg. 2020. PMID: 32519394 Free PMC article.
References
-
- Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS (2011) Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76: 301–310. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

