Microbial dysbiosis is associated with human breast cancer
- PMID: 24421902
- PMCID: PMC3885448
- DOI: 10.1371/journal.pone.0083744
Microbial dysbiosis is associated with human breast cancer
Abstract
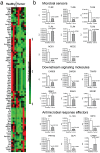
Breast cancer affects one in eight women in their lifetime. Though diet, age and genetic predisposition are established risk factors, the majority of breast cancers have unknown etiology. The human microbiota refers to the collection of microbes inhabiting the human body. Imbalance in microbial communities, or microbial dysbiosis, has been implicated in various human diseases including obesity, diabetes, and colon cancer. Therefore, we investigated the potential role of microbiota in breast cancer by next-generation sequencing using breast tumor tissue and paired normal adjacent tissue from the same patient. In a qualitative survey of the breast microbiota DNA, we found that the bacterium Methylobacterium radiotolerans is relatively enriched in tumor tissue, while the bacterium Sphingomonas yanoikuyae is relatively enriched in paired normal tissue. The relative abundances of these two bacterial species were inversely correlated in paired normal breast tissue but not in tumor tissue, indicating that dysbiosis is associated with breast cancer. Furthermore, the total bacterial DNA load was reduced in tumor versus paired normal and healthy breast tissue as determined by quantitative PCR. Interestingly, bacterial DNA load correlated inversely with advanced disease, a finding that could have broad implications in diagnosis and staging of breast cancer. Lastly, we observed lower basal levels of antibacterial response gene expression in tumor versus healthy breast tissue. Taken together, these data indicate that microbial DNA is present in the breast and that bacteria or their components may influence the local immune microenvironment. Our findings suggest a previously unrecognized link between dysbiosis and breast cancer which has potential diagnostic and therapeutic implications.
Conflict of interest statement
Figures


References
-
- Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics. CA Cancer J Clin 60: 277–300. - PubMed
-
- Madigan MP, Ziegler RG, Benichou J, Byrne C, Hoover RN (1995) Proportion of breast cancer cases in the United States explained by well-established risk factors. J Natl Cancer Inst 87: 1681–1685. - PubMed
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1131. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

