Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis
- PMID: 24422557
- PMCID: PMC3985478
- DOI: 10.1021/ja4118957
Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis
Abstract
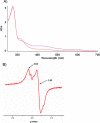
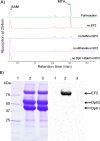
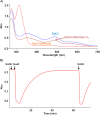
Diphthamide, the target of diphtheria toxin, is a unique posttranslational modification on translation elongation factor 2 (EF2) in archaea and eukaryotes. The biosynthesis of diphthamide was proposed to involve three steps. The first step is the transfer of the 3-amino-3-carboxypropyl group from S-adenosyl-l-methionine (SAM) to the histidine residue of EF2, forming a C-C bond. Previous genetic studies showed this step requires four proteins in eukaryotes, Dph1-Dph4. However, the exact molecular functions for the four proteins are unknown. Previous study showed that Pyrococcus horikoshii Dph2 (PhDph2), a novel iron-sulfur cluster-containing enzyme, forms a homodimer and is sufficient for the first step of diphthamide biosynthesis in vitro. Here we demonstrate by in vitro reconstitution that yeast Dph1 and Dph2 form a complex (Dph1-Dph2) that is equivalent to the homodimer of PhDph2 and is sufficient to catalyze the first step in vitro in the presence of dithionite as the reductant. We further demonstrate that yeast Dph3 (also known as KTI11), a CSL-type zinc finger protein, can bind iron and in the reduced state can serve as an electron donor to reduce the Fe-S cluster in Dph1-Dph2. Our study thus firmly establishes the functions for three of the proteins involved in eukaryotic diphthamide biosynthesis. For most radical SAM enzymes in bacteria, flavodoxins and flavodoxin reductases are believed to serve as electron donors for the Fe-S clusters. The finding that Dph3 is an electron donor for the Fe-S clusters in Dph1-Dph2 is thus interesting and opens up new avenues of research on electron transfer to Fe-S proteins in eukaryotic cells.
Figures
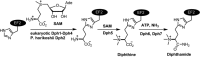
Similar articles
-
Dph3 Enables Aerobic Diphthamide Biosynthesis by Donating One Iron Atom to Transform a [3Fe-4S] to a [4Fe-4S] Cluster in Dph1-Dph2.J Am Chem Soc. 2021 Jun 30;143(25):9314-9319. doi: 10.1021/jacs.1c03956. Epub 2021 Jun 21. J Am Chem Soc. 2021. PMID: 34154323 Free PMC article.
-
The asymmetric function of Dph1-Dph2 heterodimer in diphthamide biosynthesis.J Biol Inorg Chem. 2019 Sep;24(6):777-782. doi: 10.1007/s00775-019-01702-0. Epub 2019 Aug 28. J Biol Inorg Chem. 2019. PMID: 31463593 Free PMC article.
-
Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis.Mol Biosyst. 2011 Jan;7(1):74-81. doi: 10.1039/c0mb00076k. Epub 2010 Oct 8. Mol Biosyst. 2011. PMID: 20931132 Free PMC article.
-
The diphthamide modification pathway from Saccharomyces cerevisiae--revisited.Mol Microbiol. 2014 Dec;94(6):1213-26. doi: 10.1111/mmi.12845. Epub 2014 Nov 17. Mol Microbiol. 2014. PMID: 25352115 Review.
-
Diphthamide - a conserved modification of eEF2 with clinical relevance.Trends Mol Med. 2024 Feb;30(2):164-177. doi: 10.1016/j.molmed.2023.11.008. Epub 2023 Dec 13. Trends Mol Med. 2024. PMID: 38097404 Review.
Cited by
-
Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi.Nat Struct Mol Biol. 2016 Sep;23(9):794-802. doi: 10.1038/nsmb.3265. Epub 2016 Jul 25. Nat Struct Mol Biol. 2016. PMID: 27455459 Free PMC article.
-
Cbr1 is a Dph3 reductase required for the tRNA wobble uridine modification.Nat Chem Biol. 2016 Dec;12(12):995-997. doi: 10.1038/nchembio.2190. Epub 2016 Oct 3. Nat Chem Biol. 2016. PMID: 27694803 Free PMC article.
-
DPH1 Gene Mutations Identify a Candidate SAM Pocket in Radical Enzyme Dph1•Dph2 for Diphthamide Synthesis on EF2.Biomolecules. 2023 Nov 16;13(11):1655. doi: 10.3390/biom13111655. Biomolecules. 2023. PMID: 38002337 Free PMC article.
-
Functions and Regulation of Translation Elongation Factors.Front Mol Biosci. 2022 Jan 19;8:816398. doi: 10.3389/fmolb.2021.816398. eCollection 2021. Front Mol Biosci. 2022. PMID: 35127825 Free PMC article. Review.
-
Dph3 Enables Aerobic Diphthamide Biosynthesis by Donating One Iron Atom to Transform a [3Fe-4S] to a [4Fe-4S] Cluster in Dph1-Dph2.J Am Chem Soc. 2021 Jun 30;143(25):9314-9319. doi: 10.1021/jacs.1c03956. Epub 2021 Jun 21. J Am Chem Soc. 2021. PMID: 34154323 Free PMC article.
References
-
- Robinson E. A.; Henriksen O.; Maxwell E. S. J. Biol. Chem. 1974, 249, 5088. - PubMed
-
- Van Ness B. G.; Howard J. B.; Bodley J. W. J. Biol. Chem. 1980, 255, 10710. - PubMed
-
- Van Ness B. G.; Howard J. B.; Bodley J. W. J. Biol. Chem. 1980, 255, 10717. - PubMed
-
- Collier R. J. Toxicon 2001, 39, 1793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

