HIV-1 Vif N-terminal motif is required for recruitment of Cul5 to suppress APOBEC3
- PMID: 24422669
- PMCID: PMC3937519
- DOI: 10.1186/1742-4690-11-4
HIV-1 Vif N-terminal motif is required for recruitment of Cul5 to suppress APOBEC3
Abstract
Background: HIV-1 Vif promotes the degradation of host anti-retroviral factor family, APOBEC3 proteins via the recruitment of a multi-subunit E3 ubiquitin ligase complex. The complex is composed of a scaffold protein, Cullin 5 (Cul5), RING-box protein (Rbx), a SOCS box binding protein complex, Elongins B/C (Elo B/C), as well as newly identified host co-factor, core binding factor beta (CBF-β). Cul5 has previously been shown to bind amino acids within an HCCH domain as well as a PPLP motif at the C-terminus of Vif; however, it is unclear whether Cul5 binding requires additional regions of the Vif polypeptide.
Results: Here, we provide evidence that an amino terminal region of full length Vif is necessary for the Vif-Cul5 interaction. Single alanine replacement of select amino acids spanning residues 25-30 (25VXHXMY30) reduced the ability for Vif to bind Cul5, but not CBF-β or Elo B/C in pull-down experiments. In addition, recombinant Vif mutants had a reduced binding affinity for Cul5 compared to wild-type as measured by isothermal titration calorimetry. N-terminal mutants that demonstrated reduced Cul5 binding were also unable to degrade APOBEC3G as well as APOBEC3F and were unable to restore HIV infectivity, in the presence of APOBEC3G. Although the Vif N-terminal amino acids were necessary for Cul5 interaction, the mutation of each residue to alanine induced a change in the secondary structure of the Vif-CBF-β-Elo B/C complex as suggested by results from circular dichroism spectroscopy and size-exclusion chromatography experiments. Surprisingly, the replacement of His108 to alanine also contributed to the Vif structure. Thus, it is unclear whether the amino acids contribute to a direct interaction with Cul5 or whether the amino acids are responsible for the structural organization of the Vif protein that promotes Cul5 binding.
Conclusions: Taken together, we propose a novel Vif N-terminal motif that is responsible for Vif recruitment of Cul5. Motifs in Vif that are absent from cellular proteins represent attractive targets for future HIV pharmaceutical design.
Figures


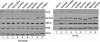



Similar articles
-
T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction.Nature. 2011 Dec 21;481(7381):376-9. doi: 10.1038/nature10718. Nature. 2011. PMID: 22190036
-
Cellular requirements for bovine immunodeficiency virus Vif-mediated inactivation of bovine APOBEC3 proteins.J Virol. 2014 Nov;88(21):12528-40. doi: 10.1128/JVI.02072-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142583 Free PMC article.
-
Requirement of HIV-1 Vif C-terminus for Vif-CBF-β interaction and assembly of CUL5-containing E3 ligase.BMC Microbiol. 2014 Nov 26;14:290. doi: 10.1186/s12866-014-0290-7. BMC Microbiol. 2014. PMID: 25424878 Free PMC article.
-
Structural insights for HIV-1 therapeutic strategies targeting Vif.Trends Biochem Sci. 2014 Sep;39(9):373-80. doi: 10.1016/j.tibs.2014.07.001. Epub 2014 Aug 12. Trends Biochem Sci. 2014. PMID: 25124760 Free PMC article. Review.
-
Antiviral roles of APOBEC proteins against HIV-1 and suppression by Vif.Arch Virol. 2009;154(10):1579-88. doi: 10.1007/s00705-009-0481-y. Epub 2009 Aug 12. Arch Virol. 2009. PMID: 19669862 Review.
Cited by
-
The ubiquitin-conjugating system: multiple roles in viral replication and infection.Cells. 2014 May 6;3(2):386-417. doi: 10.3390/cells3020386. Cells. 2014. PMID: 24805990 Free PMC article.
-
Structural perspectives on HIV-1 Vif and APOBEC3 restriction factor interactions.Protein Sci. 2020 Feb;29(2):391-406. doi: 10.1002/pro.3729. Epub 2019 Nov 29. Protein Sci. 2020. PMID: 31518043 Free PMC article. Review.
-
Mechanisms of Immune Evasion in HIV-1: The Role of Virus-Host Protein Interactions.Curr Issues Mol Biol. 2025 May 16;47(5):367. doi: 10.3390/cimb47050367. Curr Issues Mol Biol. 2025. PMID: 40699766 Free PMC article. Review.
-
Evolutionarily conserved pressure for the existence of distinct G2/M cell cycle arrest and A3H inactivation functions in HIV-1 Vif.Cell Cycle. 2015;14(6):838-47. doi: 10.1080/15384101.2014.1000212. Cell Cycle. 2015. PMID: 25590520 Free PMC article.
-
Structural Insights into APOBEC3-Mediated Lentiviral Restriction.Viruses. 2020 May 27;12(6):587. doi: 10.3390/v12060587. Viruses. 2020. PMID: 32471198 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

