A genome-wide scan for signatures of selection in Chinese indigenous and commercial pig breeds
- PMID: 24422716
- PMCID: PMC3898232
- DOI: 10.1186/1471-2156-15-7
A genome-wide scan for signatures of selection in Chinese indigenous and commercial pig breeds
Abstract
Background: Modern breeding and artificial selection play critical roles in pig domestication and shape the genetic variation of different breeds. China has many indigenous pig breeds with various characteristics in morphology and production performance that differ from those of foreign commercial pig breeds. However, the signatures of selection on genes implying for economic traits between Chinese indigenous and commercial pigs have been poorly understood.
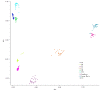
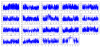
Results: We identified footprints of positive selection at the whole genome level, comprising 44,652 SNPs genotyped in six Chinese indigenous pig breeds, one developed breed and two commercial breeds. An empirical genome-wide distribution of Fst (F-statistics) was constructed based on estimations of Fst for each SNP across these nine breeds. We detected selection at the genome level using the High-Fst outlier method and found that 81 candidate genes show high evidence of positive selection. Furthermore, the results of network analyses showed that the genes that displayed evidence of positive selection were mainly involved in the development of tissues and organs, and the immune response. In addition, we calculated the pairwise Fst between Chinese indigenous and commercial breeds (CHN VS EURO) and between Northern and Southern Chinese indigenous breeds (Northern VS Southern). The IGF1R and ESR1 genes showed evidence of positive selection in the CHN VS EURO and Northern VS Southern groups, respectively.
Conclusions: In this study, we first identified the genomic regions that showed evidences of selection between Chinese indigenous and commercial pig breeds using the High-Fst outlier method. These regions were found to be involved in the development of tissues and organs, the immune response, growth and litter size. The results of this study provide new insights into understanding the genetic variation and domestication in pigs.
Figures
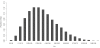


Similar articles
-
Whole-genome sequencing of European autochthonous and commercial pig breeds allows the detection of signatures of selection for adaptation of genetic resources to different breeding and production systems.Genet Sel Evol. 2020 Jun 26;52(1):33. doi: 10.1186/s12711-020-00553-7. Genet Sel Evol. 2020. PMID: 32591011 Free PMC article.
-
A genome-wide detection of selection signatures in conserved and commercial pig breeds maintained in Poland.BMC Genet. 2018 Oct 22;19(1):95. doi: 10.1186/s12863-018-0681-0. BMC Genet. 2018. PMID: 30348079 Free PMC article.
-
Genome-wide detection of selection signatures in Chinese indigenous Laiwu pigs revealed candidate genes regulating fat deposition in muscle.BMC Genet. 2018 May 18;19(1):31. doi: 10.1186/s12863-018-0622-y. BMC Genet. 2018. PMID: 29776331 Free PMC article.
-
Genetic resources, genome mapping and evolutionary genomics of the pig (Sus scrofa).Int J Biol Sci. 2007 Feb 10;3(3):153-65. doi: 10.7150/ijbs.3.153. Int J Biol Sci. 2007. PMID: 17384734 Free PMC article. Review.
-
Assembly of novel sequences for Chinese domestic pigs reveals new genes and regulatory variants providing new insights into their diversity.Genomics. 2024 Mar;116(2):110782. doi: 10.1016/j.ygeno.2024.110782. Epub 2024 Jan 3. Genomics. 2024. PMID: 38176574 Review.
Cited by
-
Whole-genome sequencing of European autochthonous and commercial pig breeds allows the detection of signatures of selection for adaptation of genetic resources to different breeding and production systems.Genet Sel Evol. 2020 Jun 26;52(1):33. doi: 10.1186/s12711-020-00553-7. Genet Sel Evol. 2020. PMID: 32591011 Free PMC article.
-
Assessment of Heterozygosity and Genome-Wide Analysis of Heterozygosity Regions in Two Duroc Pig Populations.Front Genet. 2022 Jan 27;12:812456. doi: 10.3389/fgene.2021.812456. eCollection 2021. Front Genet. 2022. PMID: 35154256 Free PMC article.
-
Detection of genomic structural variations in Guizhou indigenous pigs and the comparison with other breeds.PLoS One. 2018 Mar 20;13(3):e0194282. doi: 10.1371/journal.pone.0194282. eCollection 2018. PLoS One. 2018. PMID: 29558483 Free PMC article.
-
Small Intestinal Digestive Functions and Feed Efficiency Differ in Different Pig Breeds.Animals (Basel). 2023 Mar 26;13(7):1172. doi: 10.3390/ani13071172. Animals (Basel). 2023. PMID: 37048428 Free PMC article.
-
Integrating spatial transcriptomics and single-nucleus RNA-seq revealed the specific inhibitory effects of TGF-β on intramuscular fat deposition.Sci China Life Sci. 2025 Mar;68(3):746-763. doi: 10.1007/s11427-024-2696-5. Epub 2024 Oct 12. Sci China Life Sci. 2025. PMID: 39422812
References
-
- Albarella U, Dobney K, Ervynk A, Rowley-Conwy P. Pigs and Humans: 10,000 Years of Interaction. Oxford: Oxford University Press; 2007.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

