Correction: Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi
- PMID: 24422981
- PMCID: PMC3893384
- DOI: 10.1186/1471-2164-15-6
Correction: Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi
Abstract
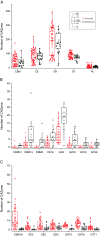
The version of this article published in BMC Genomics 2013, 14: 274, contains 9 unpublished genomes (Botryobasidium botryosum, Gymnopus luxurians, Hypholoma sublateritium, Jaapia argillacea, Hebeloma cylindrosporum, Conidiobolus coronatus, Laccaria amethystina, Paxillus involutus, and P. rubicundulus) downloaded from JGI website. In this correction, we removed these genomes after discussion with editors and data producers whom we should have contacted before downloading these genomes. Removing these data did not alter the principle results and conclusions of our original work. The relevant Figures 1, 2, 3, 4 and 6; and Table 1 have been revised. Additional files 1, 3, 4, and 5 were also revised. We would like to apologize for any confusion or inconvenience this may have caused.
Background: Fungi produce a variety of carbohydrate activity enzymes (CAZymes) for the degradation of plant polysaccharide materials to facilitate infection and/or gain nutrition. Identifying and comparing CAZymes from fungi with different nutritional modes or infection mechanisms may provide information for better understanding of their life styles and infection models. To date, over hundreds of fungal genomes are publicly available. However, a systematic comparative analysis of fungal CAZymes across the entire fungal kingdom has not been reported.
Results: In this study, we systemically identified glycoside hydrolases (GHs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), and glycosyltransferases (GTs) as well as carbohydrate-binding modules (CBMs) in the predicted proteomes of 94 representative fungi from Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota. Comparative analysis of these CAZymes that play major roles in plant polysaccharide degradation revealed that fungi exhibit tremendous diversity in the number and variety of CAZymes. Among them, some families of GHs and CEs are the most prevalent CAZymes that are distributed in all of the fungi analyzed. Importantly, cellulases of some GH families are present in fungi that are not known to have cellulose-degrading ability. In addition, our results also showed that in general, plant pathogenic fungi have the highest number of CAZymes. Biotrophic fungi tend to have fewer CAZymes than necrotrophic and hemibiotrophic fungi. Pathogens of dicots often contain more pectinases than fungi infecting monocots. Interestingly, besides yeasts, many saprophytic fungi that are highly active in degrading plant biomass contain fewer CAZymes than plant pathogenic fungi. Furthermore, analysis of the gene expression profile of the wheat scab fungus Fusarium graminearum revealed that most of the CAZyme genes related to cell wall degradation were up-regulated during plant infection. Phylogenetic analysis also revealed a complex history of lineage-specific expansions and attritions for the PL1 family.
Conclusions: Our study provides insights into the variety and expansion of fungal CAZyme classes and revealed the relationship of CAZyme size and diversity with their nutritional strategy and host specificity.
Figures



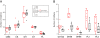
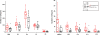

Erratum for
-
Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi.BMC Genomics. 2013 Apr 23;14:274. doi: 10.1186/1471-2164-14-274. BMC Genomics. 2013. PMID: 23617724 Free PMC article.
Similar articles
-
Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi.BMC Genomics. 2013 Apr 23;14:274. doi: 10.1186/1471-2164-14-274. BMC Genomics. 2013. PMID: 23617724 Free PMC article.
-
Insight into plant cell wall degradation and pathogenesis of Ganoderma boninense via comparative genome analysis.PeerJ. 2019 Dec 18;7:e8065. doi: 10.7717/peerj.8065. eCollection 2019. PeerJ. 2019. PMID: 31879570 Free PMC article.
-
Carbohydrate-active enzymes in Trichoderma harzianum: a bioinformatic analysis bioprospecting for key enzymes for the biofuels industry.BMC Genomics. 2017 Oct 12;18(1):779. doi: 10.1186/s12864-017-4181-9. BMC Genomics. 2017. PMID: 29025413 Free PMC article.
-
Whole-Genome Sequencing and Comparative Genomics Analysis of the Wild Edible Mushroom (Gomphus purpuraceus) Provide Insights into Its Potential Food Application and Artificial Domestication.Genes (Basel). 2022 Sep 10;13(9):1628. doi: 10.3390/genes13091628. Genes (Basel). 2022. PMID: 36140797 Free PMC article. Review.
-
Modulating Transcriptional Regulation of Plant Biomass Degrading Enzyme Networks for Rational Design of Industrial Fungal Strains.Front Bioeng Biotechnol. 2018 Sep 25;6:133. doi: 10.3389/fbioe.2018.00133. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30320082 Free PMC article. Review.
Cited by
-
The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases.Sci Rep. 2015 Jun 18;5:11153. doi: 10.1038/srep11153. Sci Rep. 2015. PMID: 26084520 Free PMC article.
-
Biotechnology Potential of Marine Fungi Degrading Plant and Algae Polymeric Substrates.Front Microbiol. 2018 Jul 10;9:1527. doi: 10.3389/fmicb.2018.01527. eCollection 2018. Front Microbiol. 2018. PMID: 30050513 Free PMC article. Review.
-
De Novo Long-Read Whole-Genome Assemblies and the Comparative Pan-Genome Analysis of Ascochyta Blight Pathogens Affecting Field Pea.J Fungi (Basel). 2022 Aug 22;8(8):884. doi: 10.3390/jof8080884. J Fungi (Basel). 2022. PMID: 36012871 Free PMC article.
-
Deciphering the Infectious Process of Colletotrichum lupini in Lupin through Transcriptomic and Proteomic Analysis.Microorganisms. 2020 Oct 21;8(10):1621. doi: 10.3390/microorganisms8101621. Microorganisms. 2020. PMID: 33096724 Free PMC article.
-
Simultaneous Silencing of Xylanase Genes in Botrytis cinerea.Front Plant Sci. 2017 Dec 22;8:2174. doi: 10.3389/fpls.2017.02174. eCollection 2017. Front Plant Sci. 2017. PMID: 29312413 Free PMC article.
References
-
- Couturier M, Navarro D, Olive C, Chevret D, Haon M, Favel A, Lesage-Meessen L, Henrissat B, Coutinho PM, Berrin JG. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genomics. 2012;15:57. doi: 10.1186/1471-2164-13-57. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

