AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation
- PMID: 24423872
- PMCID: PMC3973335
- DOI: 10.1093/nar/gkt1379
AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation
Abstract
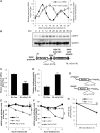
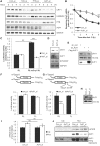
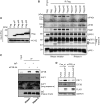
In the present study, we investigated the 3' untranslated region (UTR) of the mouse core clock gene cryptochrome 1 (Cry1) at the post-transcriptional level, particularly its translational regulation. Interestingly, the 3'UTR of Cry1 mRNA decreased its mRNA levels but increased protein amounts. The 3'UTR is widely known to function as a cis-acting element of mRNA degradation. The 3'UTR also provides a binding site for microRNA and mainly suppresses translation of target mRNAs. We found that AU-rich element RNA binding protein 1 (AUF1) directly binds to the Cry1 3'UTR and regulates translation of Cry1 mRNA. AUF1 interacted with eukaryotic translation initiation factor 3 subunit B and also directly associated with ribosomal protein S3 or ribosomal protein S14, resulting in translation of Cry1 mRNA in a 3'UTR-dependent manner. Expression of cytoplasmic AUF1 and binding of AUF1 to the Cry1 3'UTR were parallel to the circadian CRY1 protein profile. Our results suggest that the 3'UTR of Cry1 is important for its rhythmic translation, and AUF1 bound to the 3'UTR facilitates interaction with the 5' end of mRNA by interacting with translation initiation factors and recruiting the 40S ribosomal subunit to initiate translation of Cry1 mRNA.
Figures




Similar articles
-
Interaction between 3' untranslated region of calcitonin receptor messenger ribonucleic acid (RNA) and adenylate/uridylate (AU)-rich element binding proteins (AU-rich RNA-binding factor 1 and Hu antigen R).Endocrinology. 2004 Apr;145(4):1730-8. doi: 10.1210/en.2003-0862. Epub 2004 Jan 8. Endocrinology. 2004. PMID: 14715706
-
Short-lived AUF1 p42-binding mRNAs of RANKL and BCL6 have two distinct instability elements each.PLoS One. 2018 Nov 12;13(11):e0206823. doi: 10.1371/journal.pone.0206823. eCollection 2018. PLoS One. 2018. PMID: 30418981 Free PMC article.
-
Post-transcriptional control of the MCT-1-associated protein DENR/DRP by RNA-binding protein AUF1.Cancer Genomics Proteomics. 2007 May-Jun;4(3):233-9. Cancer Genomics Proteomics. 2007. PMID: 17878526
-
Physiological networks and disease functions of RNA-binding protein AUF1.Wiley Interdiscip Rev RNA. 2014 Jul-Aug;5(4):549-64. doi: 10.1002/wrna.1230. Epub 2014 Mar 28. Wiley Interdiscip Rev RNA. 2014. PMID: 24687816 Review.
-
The role of AUF1 in regulated mRNA decay.Wiley Interdiscip Rev RNA. 2010 Nov-Dec;1(3):457-73. doi: 10.1002/wrna.26. Wiley Interdiscip Rev RNA. 2010. PMID: 21956942 Free PMC article. Review.
Cited by
-
HnRNP Q Has a Suppressive Role in the Translation of Mouse Cryptochrome1.PLoS One. 2016 Jul 8;11(7):e0159018. doi: 10.1371/journal.pone.0159018. eCollection 2016. PLoS One. 2016. PMID: 27392095 Free PMC article.
-
Functional analysis of a novel cryptochrome gene (GbCRY1) from Ginkgo biloba.Plant Signal Behav. 2021 Feb 1;16(2):1850627. doi: 10.1080/15592324.2020.1850627. Epub 2020 Dec 1. Plant Signal Behav. 2021. PMID: 33258712 Free PMC article.
-
Heterogeneous nuclear ribonucleoprotein A1 regulates rhythmic synthesis of mouse Nfil3 protein via IRES-mediated translation.Sci Rep. 2017 Feb 21;7:42882. doi: 10.1038/srep42882. Sci Rep. 2017. PMID: 28220845 Free PMC article.
-
The molecular clockwork of mammalian cells.Semin Cell Dev Biol. 2022 Jun;126:87-96. doi: 10.1016/j.semcdb.2021.03.012. Epub 2021 Mar 31. Semin Cell Dev Biol. 2022. PMID: 33810978 Free PMC article. Review.
-
The Biological Roles of Translation Initiation Factor 3b.Int J Biol Sci. 2018 Sep 7;14(12):1630-1635. doi: 10.7150/ijbs.26932. eCollection 2018. Int J Biol Sci. 2018. PMID: 30416377 Free PMC article. Review.
References
-
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

