Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis
- PMID: 24423873
- PMCID: PMC3973336
- DOI: 10.1093/nar/gkt1376
Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis
Abstract
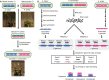
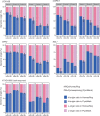
Genome duplication with hybridization, or allopolyploidization, occurs commonly in plants, and is considered to be a strong force for generating new species. However, genome-wide quantification of homeolog expression ratios was technically hindered because of the high homology between homeologous gene pairs. To quantify the homeolog expression ratio using RNA-seq obtained from polyploids, a new method named HomeoRoq was developed, in which the genomic origin of sequencing reads was estimated using mismatches between the read and each parental genome. To verify this method, we first assembled the two diploid parental genomes of Arabidopsis halleri subsp. gemmifera and Arabidopsis lyrata subsp. petraea (Arabidopsis petraea subsp. umbrosa), then generated a synthetic allotetraploid, mimicking the natural allopolyploid Arabidopsis kamchatica. The quantified ratios corresponded well to those obtained by Pyrosequencing. We found that the ratios of homeologs before and after cold stress treatment were highly correlated (r = 0.870). This highlights the presence of nonstochastic polyploid gene regulation despite previous research identifying stochastic variation in expression. Moreover, our new statistical test incorporating overdispersion identified 226 homeologs (1.11% of 20 369 expressed homeologs) with significant ratio changes, many of which were related to stress responses. HomeoRoq would contribute to the study of the genes responsible for polyploid-specific environmental responses.
Figures



References
-
- Jiao YN, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang HY, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–U113. - PubMed
-
- Stebbins GL. Variation and Evolution in Plants. New York: Columbia University; 1950.
-
- Stebbins GL. Chromosomal evolution in Higher Plants. London: Edward Arnold; 1971.
-
- Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

