Leptin acts independently of food intake to modulate gut microbial composition in male mice
- PMID: 24424041
- PMCID: PMC3929727
- DOI: 10.1210/en.2013-1085
Leptin acts independently of food intake to modulate gut microbial composition in male mice
Abstract
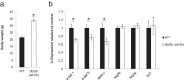
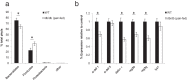
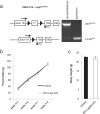
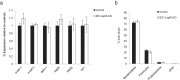
Shifts in the composition of gut bacterial populations can alter host metabolism and may contribute to the pathogenesis of metabolic disorders, including obesity. Mice deficient in leptin action are obese with altered microbiota and increased susceptibility to certain intestinal pathogens. Because antimicrobial peptides (AMPs) secreted by Paneth cells represent a major mechanism by which the host influences the gut microbiome, we examined the mRNA expression of gut AMPs, several of which were decreased in leptin receptor (LepR)-deficient db/db mice, suggesting a potential role for AMP modulation of microbiota composition. To address the extent to which the alterations in gut microbiota and AMP mRNA expression in db/db mice result from increased food intake vs other defects in leptin action, we examined the effects of pair feeding and gut epithelial LepRb ablation on AMP mRNA expression and microbiota composition. We found that the phylum-level changes in fecal microbial content and AMP gene expression persist in pair-fed db/db mice, suggesting that these differences do not stem from hyperphagia alone. In addition, despite recent evidence to support a role for intestinal epithelial LepRb signaling in pathogen susceptibility, ablation of LepRb from the intestinal epithelium fails to alter body weight, composition of the microbiota, or AMP expression, suggesting a role for LepRb elsewhere for this regulation. Indeed, gut LepRb cells are not epithelial but rather constitute a previously uncharacterized population of perivascular cells within the intestinal submucosa. Overall, our data reveal a role for LepRb signaling extrinsic to the intestinal epithelium and independent of food intake in the control of the gut microbiome.
Figures

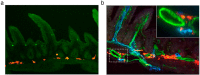
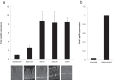

Comment in
-
Old dog, new trick: a direct role for leptin in regulating microbiota composition.Endocrinology. 2014 Mar;155(3):653-5. doi: 10.1210/en.2014-1078. Endocrinology. 2014. PMID: 24564414 Free PMC article. No abstract available.
References
-
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

